Proteomic and phosphoproteomic characterization of cardiovascular tissues after long term exposure to simulated space radiation
- PMID: 38699144
- PMCID: PMC11063234
- DOI: 10.3389/fphys.2024.1248276
Proteomic and phosphoproteomic characterization of cardiovascular tissues after long term exposure to simulated space radiation
Abstract
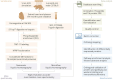
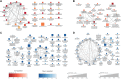
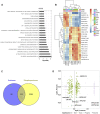
Introduction: It may take decades to develop cardiovascular dysfunction following exposure to high doses of ionizing radiation from medical therapy or from nuclear accidents. Since astronauts may be exposed continually to a complex space radiation environment unlike that experienced on Earth, it is unresolved whether there is a risk to cardiovascular health during long-term space exploration missions. Previously, we have described that mice exposed to a single dose of simplified Galactic Cosmic Ray (GCR5-ion) develop cardiovascular dysfunction by 12 months post-radiation. Methods: To investigate the biological basis of this dysfunction, here we performed a quantitative mass spectrometry-based proteomics analysis of heart tissue (proteome and phosphoproteome) and plasma (proteome only) from these mice at 8 months post-radiation. Results: Differentially expressed proteins (DEPs) for irradiated versus sham irradiated samples (fold-change ≥1.2 and an adjusted p-value of ≤0.05) were identified for each proteomics data set. For the heart proteome, there were 87 significant DEPs (11 upregulated and 76 downregulated); for the heart phosphoproteome, there were 60 significant differentially phosphorylated peptides (17 upregulated and 43 downregulated); and for the plasma proteome, there was only one upregulated protein. A Gene Set Enrichment Analysis (GSEA) technique that assesses canonical pathways from BIOCARTA, KEGG, PID, REACTOME, and WikiPathways revealed significant perturbation in pathways in each data set. For the heart proteome, 166 pathways were significantly altered (36 upregulated and 130 downregulated); for the plasma proteome, there were 73 pathways significantly altered (25 upregulated and 48 downregulated); and for the phosphoproteome, there were 223 pathways significantly affected at 0.1 adjusted p-value cutoff. Pathways related to inflammation were the most highly perturbed in the heart and plasma. In line with sustained inflammation, neutrophil extracellular traps (NETs) were demonstrated to be increased in GCR5-ion irradiated hearts at 12-month post irradiation. NETs play a fundamental role in combating bacterial pathogens, modulating inflammatory responses, inflicting damage on healthy tissues, and escalating vascular thrombosis. Discussion: These findings suggest that a single exposure to GCR5-ion results in long-lasting changes in the proteome and that these proteomic changes can potentiate acute and chronic health issues for astronauts, such as what we have previously described with late cardiac dysfunction in these mice.
Keywords: cardiovascular degeneration; cardiovascular disease; galactic cosmic ray; ionizing radiation; mass spectrometry; phosphoproteomics; proteomics; space radiation.
Copyright © 2024 Kidane, Lee, Smith, Wang, Mirza, Sharma, Lobo, Dewan, Chen, Diaz, Pla, Foster and Bowles.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Amino M., Yoshioka K., Fujibayashi D., Hashida T., Furusawa Y., Zareba W., et al. (2010). Year-long upregulation of connexin43 in rabbit hearts by heavy ion irradiation. Am. J. Physiol. Heart Circ. Physiol. 298 (3), H1014–H1021. Epub 2010 Jan 8. PMID: 20061548. 10.1152/ajpheart.00160.2009 - DOI - PubMed
-
- Azimzadeh O., Moertl S., Ramadan R., Baselet B., Laiakis E. C., Sebastian S., et al. (2022). Application of radiation omics in the development of adverse outcome pathway networks: an example of radiation induced cardiovascular disease. Int. J. Radiat. Biol. 98 (12), 1722–1751. 10.1080/09553002.2022.2110325 - DOI - PubMed
-
- Beghi S., Furmanik M., Jaminon A., Veltrop R., Rapp N., Wichapong K., et al. (2022). Calcium signalling in heart and vessels: role of calmodulin and downstream calmodulin-dependent protein kinases. Int. J. Mol. Sci. 23 (24), 16139. PMID: 36555778; PMCID: PMC9783221. 10.3390/ijms232416139 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

