Streamlining the automated discovery of porous organic cages
- PMID: 38699265
- PMCID: PMC11062116
- DOI: 10.1039/d3sc06133g
Streamlining the automated discovery of porous organic cages
Abstract
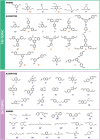
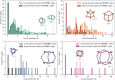
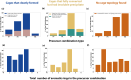
Self-assembly through dynamic covalent chemistry (DCC) can yield a range of multi-component organic assemblies. The reversibility and dynamic nature of DCC has made prediction of reaction outcome particularly difficult and thus slows the discovery rate of new organic materials. In addition, traditional experimental processes are time-consuming and often rely on serendipity. Here, we present a streamlined hybrid workflow that combines automated high-throughput experimentation, automated data analysis, and computational modelling, to accelerate the discovery process of one particular subclass of molecular organic materials, porous organic cages. We demonstrate how the design and implementation of this workflow aids in the identification of organic cages with desirable properties. The curation of a precursor library of 55 tri- and di-topic aldehyde and amine precursors enabled the experimental screening of 366 imine condensation reactions experimentally, and 1464 hypothetical organic cage outcomes to be computationally modelled. From the screen, 225 cages were identified experimentally using mass spectrometry, 54 of which were cleanly formed as a single topology as determined by both turbidity measurements and 1H NMR spectroscopy. Integration of these characterisation methods into a fully automated Python pipeline, named cagey, led to over a 350-fold decrease in the time required for data analysis. This work highlights the advantages of combining automated synthesis, characterisation, and analysis, for large-scale data curation towards an accessible data-driven materials discovery approach.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Stach E. DeCost B. Kusne A. G. Hattrick-Simpers J. Brown K. A. Reyes K. G. Schrier J. Billinge S. Buonassisi T. Foster I. Gomes C. P. Gregoire J. M. Mehta A. Montoya J. Olivetti E. Park C. Rotenberg E. Saikin S. K. Smullin S. Stanev V. Maruyama B. Matter. 2021;4:2702–2726. doi: 10.1016/j.matt.2021.06.036. - DOI
-
- Little M. A. Cooper A. I. Adv. Funct. Mater. 2020;30:1909842. doi: 10.1002/adfm.201909842. - DOI
LinkOut - more resources
Full Text Sources

