This is a preprint.
Longitudinal cervicovaginal microbiome and virome alterations during ART and discordant shedding in women living with HIV
- PMID: 38699319
- PMCID: PMC11065064
- DOI: 10.21203/rs.3.rs-4078561/v1
Longitudinal cervicovaginal microbiome and virome alterations during ART and discordant shedding in women living with HIV
Abstract
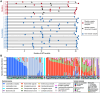
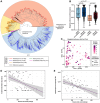
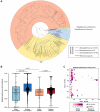
Despite successful suppression of plasma HIV replication by antiretroviral therapy (ART), some women living with HIV (WLHIV) can still experience genital HIV shedding (discordant shedding). Female genital tract (FGT) microbiome and virome dynamics during long-term ART in WLHIV are poorly understood but might contribute to discordant HIV shedding, as the microbiome and virome are known to influence FGT health. To understand FGT microbial communities over time during ART usage and discordant shedding, we characterized the microbiome and virome in 125 cervicovaginal specimens collected over two years in 31 WLHIV in Lima, Peru. Intrapersonal bacterial microbiome variation was higher in HIV shedders compared to non-shedders. Cervicovaginal virome composition changed over time, particularly in non-shedders. Specifically, anellovirus relative abundance was inversely associated with ART duration and CD4 counts. Our results suggest that discordant HIV shedding is associated with FGT microbiome instability, and immune recovery during ART influences FGT virome composition.
Figures


Similar articles
-
Longitudinal cervicovaginal bacteriome and virome alterations associate with discordant shedding and ART duration in women living with HIV in Peru.Nat Commun. 2025 Aug 25;16(1):7904. doi: 10.1038/s41467-025-63158-y. Nat Commun. 2025. PMID: 40854874
-
Breast Milk Virome and Bacterial Microbiome Resilience in Kenyan Women Living with HIV.mSystems. 2021 Mar 16;6(2):e01079-20. doi: 10.1128/mSystems.01079-20. mSystems. 2021. PMID: 33727396 Free PMC article.
-
Cervicovaginal DNA Virome Alterations Are Associated with Genital Inflammation and Microbiota Composition.mSystems. 2022 Apr 26;7(2):e0006422. doi: 10.1128/msystems.00064-22. Epub 2022 Mar 28. mSystems. 2022. PMID: 35343798 Free PMC article.
-
The impact of the female genital tract microbiome in women health and reproduction: a review.J Assist Reprod Genet. 2021 Oct;38(10):2519-2541. doi: 10.1007/s10815-021-02247-5. Epub 2021 Jun 10. J Assist Reprod Genet. 2021. PMID: 34110573 Free PMC article. Review.
-
Adverse perinatal outcomes associated with antiretroviral therapy in women living with HIV: A systematic review and meta-analysis.Front Med (Lausanne). 2023 Feb 3;9:924593. doi: 10.3389/fmed.2022.924593. eCollection 2022. Front Med (Lausanne). 2023. PMID: 36816720 Free PMC article.
References
-
- Joint United Nations Programme on HIV and AIDS. HIV estimates with uncertainty bounds 1990-Present.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

