Loss-of-function OGFRL1 variants identified in autosomal recessive cherubism families
- PMID: 38699440
- PMCID: PMC11062026
- DOI: 10.1093/jbmrpl/ziae050
Loss-of-function OGFRL1 variants identified in autosomal recessive cherubism families
Abstract
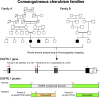
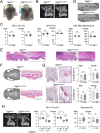
Cherubism (OMIM 118400) is a rare craniofacial disorder in children characterized by destructive jawbone expansion due to the growth of inflammatory fibrous lesions. Our previous studies have shown that gain-of-function mutations in SH3 domain-binding protein 2 (SH3BP2) are responsible for cherubism and that a knock-in mouse model for cherubism recapitulates the features of cherubism, such as increased osteoclast formation and jawbone destruction. To date, SH3BP2 is the only gene identified to be responsible for cherubism. Since not all patients clinically diagnosed with cherubism had mutations in SH3BP2, we hypothesized that there may be novel cherubism genes and that these genes may play a role in jawbone homeostasis. Here, using whole exome sequencing, we identified homozygous loss-of-function variants in the opioid growth factor receptor like 1 (OGFRL1) gene in 2 independent autosomal recessive cherubism families from Syria and India. The newly identified pathogenic homozygous variants were not reported in any variant databases, suggesting that OGFRL1 is a novel gene responsible for cherubism. Single cell analysis of mouse jawbone tissue revealed that Ogfrl1 is highly expressed in myeloid lineage cells. We generated OGFRL1 knockout mice and mice carrying the Syrian frameshift mutation to understand the in vivo role of OGFRL1. However, neither mouse model recapitulated human cherubism or the phenotypes exhibited by SH3BP2 cherubism mice under physiological and periodontitis conditions. Unlike bone marrow-derived M-CSF-dependent macrophages (BMMs) carrying the SH3BP2 cherubism mutation, BMMs lacking OGFRL1 or carrying the Syrian mutation showed no difference in TNF-ɑ mRNA induction by LPS or TNF-ɑ compared to WT BMMs. Osteoclast formation induced by RANKL was also comparable. These results suggest that the loss-of-function effects of OGFRL1 in humans differ from those in mice and highlight the fact that mice are not always an ideal model for studying rare craniofacial bone disorders.
Keywords: OGFRL1; autosomal recessive; cherubism; loss of function; mouse model; mutation; rare disease; whole exome sequencing.
© The Author(s) 2024. Published by Oxford University Press on behalf of the American Society for Bone and Mineral Research.
Conflict of interest statement
All authors state that they have no conflicts of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

