Role of MIC levels and 23S rRNA mutation sites to clarithromycin in 14-day clarithromycin bismuth quadruple therapy for Helicobacter pylori eradication: A prospective trial in Beijing
- PMID: 38699713
- PMCID: PMC11063421
- DOI: 10.1016/j.heliyon.2024.e29774
Role of MIC levels and 23S rRNA mutation sites to clarithromycin in 14-day clarithromycin bismuth quadruple therapy for Helicobacter pylori eradication: A prospective trial in Beijing
Abstract
Background: Rising clarithromycin resistance undermines Helicobacter pylori (H. pylori) treatment efficacy. We aimed to determine clarithromycin's minimum inhibitory concentration (MIC) levels and identify specific mutation sites in the 23S ribosomal subunit (23S rRNA) that predict treatment outcomes in a 14-day regimen of clarithromycin bismuth quadruple therapy (amoxicillin 1g, clarithromycin 500 mg, rabeprazole 10 mg, and colloidal bismuth pectin 200 mg).
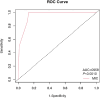
Materials and methods: We included adult H. pylori patients who hadn't previously undergone clarithromycin-based treatment, either as initial or rescue therapy. Exclusions were made for penicillin allergy, recent use of related medications, severe illnesses, or inability to cooperate. Patients underwent a 14-day clarithromycin bismuth quadruple therapy. Gastric mucosa specimens were obtained during endoscopy before eradication. MIC against amoxicillin and clarithromycin was determined using the E-test method. The receiver operating characteristic (ROC) curve helped to find the optimal clarithromycin resistance MIC breakpoint. Genetic sequences of H. pylori 23S rRNA were identified through Sanger Sequencing. (ChiCTR2200061476).
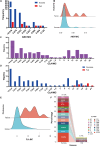
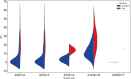
Results: Out of 196 patients recruited, 92 met the inclusion criteria for the per-protocol (PP) population. The overall intention-to-treat (ITT) eradication rate was 80.00 % (84/105), while the modified intention-to-treat (MITT) and PP eradication rates were 90.32 % (84/93) and 91.30 % (84/92) respectively. No amoxicillin resistance was observed, but clarithromycin resistance rates were 36.19 % (38/105), 35.48 % (33/93), and 34.78 % (33/92) in the ITT, MITT, and PP populations respectively. Compared with the traditional clarithromycin resistance breakpoint of 0.25 μg/mL, a MIC threshold of 12 μg/mL predicted better eradication. Among 173 mutations on 152 sites in the 23S rRNA gene, only the 2143A > G mutation could predict eradication outcomes (p < 0.000).
Conclusions: Interpretation of elevated MIC values is crucial in susceptibility testing, rather than a binary "susceptible" or "resistant" classification. The 2143A > G mutation has limited specificity in predicting eradication outcomes, necessitating further investigation into additional mutation sites associated with clarithromycin resistance.
Keywords: 23S rRNA mutation sites; Antibiotic resistance; Clarithromycin; Helicobacter pylori; Minimum inhibitory concentration (MIC); Receiver operating characteristic (ROC).
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Comparison of 10 and 14 days of antofloxacin-based versus 14 days of clarithromycin-based bismuth quadruple therapy for Helicobacter pylori eradication: A randomized trial.Clin Res Hepatol Gastroenterol. 2023 Jan;47(1):102052. doi: 10.1016/j.clinre.2022.102052. Epub 2022 Nov 15. Clin Res Hepatol Gastroenterol. 2023. PMID: 36400418 Clinical Trial.
-
A randomized controlled trial to compare Helicobacter pylori eradication rates between the empirical concomitant therapy and tailored therapy based on 23S rRNA point mutations.Medicine (Baltimore). 2022 Aug 19;101(33):e30069. doi: 10.1097/MD.0000000000030069. Medicine (Baltimore). 2022. PMID: 35984159 Free PMC article. Clinical Trial.
-
Optimizing the MIC breakpoints of amoxicillin and tetracycline for antibiotic selection in the rescue therapy of H. pylori with bismuth quadruple regimen.Eur J Clin Pharmacol. 2020 Nov;76(11):1581-1589. doi: 10.1007/s00228-020-02938-5. Epub 2020 Jun 27. Eur J Clin Pharmacol. 2020. PMID: 32591943
-
Molecular testing-guided therapy versus susceptibility testing-guided therapy in first-line and third-line Helicobacter pylori eradication: two multicentre, open-label, randomised controlled, non-inferiority trials.Lancet Gastroenterol Hepatol. 2023 Jul;8(7):623-634. doi: 10.1016/S2468-1253(23)00097-3. Epub 2023 May 10. Lancet Gastroenterol Hepatol. 2023. PMID: 37178702 Clinical Trial.
-
The Challenges of Treating a Helicobacter pylori Infection following the COVID-19 Pandemic in Croatia: A Review.J Clin Med. 2024 Sep 27;13(19):5762. doi: 10.3390/jcm13195762. J Clin Med. 2024. PMID: 39407822 Free PMC article. Review.
Cited by
-
Dynamic interaction of antibiotic resistance between plant microbiome and organic fertilizers: sources, dissemination, and health risks.World J Microbiol Biotechnol. 2024 Dec 18;41(1):4. doi: 10.1007/s11274-024-04214-5. World J Microbiol Biotechnol. 2024. PMID: 39690351 Review.
References
-
- Ren S., Cai P., Liu Y., et al. Prevalence of Helicobacter pylori infection in China: a systematic review and meta-analysis. J gastroen hepatol. 2022;37(3):464–470. - PubMed
-
- Malfertheiner P., Megraud F., Rokkas T., et al. Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report. Gut. 2022 gutjnl-2022-327745. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

