Genomic and phenotypic characterization of Mycobacterium tuberculosis' closest-related non-tuberculous mycobacteria
- PMID: 38700329
- PMCID: PMC11237670
- DOI: 10.1128/spectrum.04126-23
Genomic and phenotypic characterization of Mycobacterium tuberculosis' closest-related non-tuberculous mycobacteria
Abstract
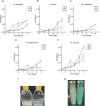
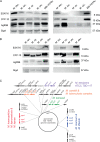
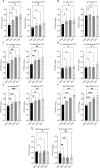
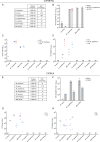
Four species of non-tuberculous mycobacteria (NTM) rated as biosafety level 1 or 2 (BSL-1/BSL-2) organisms and showing higher genomic similarity with Mycobacterium tuberculosis (Mtb) than previous comparator species Mycobacterium kansasii and Mycobacterium marinum were subjected to genomic and phenotypic characterization. These species named Mycobacterium decipiens, Mycobacterium lacus, Mycobacterium riyadhense, and Mycobacterium shinjukuense might represent "missing links" between low-virulent mycobacterial opportunists and the highly virulent obligate pathogen Mtb. We confirmed that M. decipiens is the closest NTM species to Mtb currently known and found that it has an optimal growth temperature of 32°C-35°C and not 37°C. M. decipiens showed resistance to rifampicin, isoniazid, and ethambutol, whereas M. lacus and M. riyadhense showed resistance to isoniazid and ethambutol. M. shinjukuense was sensitive to all three first-line TB drugs, and all four species were sensitive to bedaquiline, a third-generation anti-TB drug. Our results suggest these four NTM may be useful models for the identification and study of new anti-TB molecules, facilitated by their culture under non-BSL-3 conditions as compared to Mtb. M. riyadhense was the most virulent of the four species in cellular and mouse infection models. M. decipiens also multiplied in THP-1 cells at 35°C but was growth impaired at 37°C. Genomic comparisons showed that the espACD locus, essential for the secretion of ESX-1 proteins in Mtb, was present only in M. decipiens, which was able to secrete ESAT-6 and CFP-10, whereas secretion of these antigens varied in the other species, making the four species interesting examples for studying ESX-1 secretion mechanisms.IMPORTANCEIn this work, we investigated recently identified opportunistic mycobacterial pathogens that are genomically more closely related to Mycobacterium tuberculosis (Mtb) than previously used comparator species Mycobacterium kansasii and Mycobacterium marinum. We confirmed that Mycobacterium decipiens is the currently closest known species to the tubercle bacilli, represented by Mycobacterium canettii and Mtb strains. Surprisingly, the reference strain of Mycobacterium riyadhense (DSM 45176), which was purchased as a biosafety level 1 (BSL-1)-rated organism, was the most virulent of the four species in the tested cellular and mouse infection models, suggesting that a BSL-2 rating might be more appropriate for this strain than the current BSL-1 rating. Our work establishes the four NTM species as interesting study models to obtain new insights into the evolutionary mechanisms and phenotypic particularities of mycobacterial pathogens that likely have also impacted the evolution of the key pathogen Mtb.
Keywords: ESX-1; Mycobacterium tuberculosis; evolution; mycobacterial model; phylotype; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Evaluation of the STANDARD M10 MDR-TB and MTB/NTM assays for the detection of Mycobacterium tuberculosis, rifampicin and isoniazid resistance, and nontuberculous mycobacteria in a low-incidence setting.J Clin Microbiol. 2024 Oct 16;62(10):e0040224. doi: 10.1128/jcm.00402-24. Epub 2024 Sep 19. J Clin Microbiol. 2024. PMID: 39297626 Free PMC article.
-
Shared Pathogenomic Patterns Characterize a New Phylotype, Revealing Transition toward Host-Adaptation Long before Speciation of Mycobacterium tuberculosis.Genome Biol Evol. 2019 Aug 1;11(8):2420-2438. doi: 10.1093/gbe/evz162. Genome Biol Evol. 2019. PMID: 31368488 Free PMC article.
-
Non-tuberculous Mycobacterium species causing mycobacteriosis in farmed aquatic animals of South Africa.BMC Microbiol. 2018 Apr 13;18(1):32. doi: 10.1186/s12866-018-1177-9. BMC Microbiol. 2018. PMID: 29653505 Free PMC article.
-
Of tuberculosis and non-tuberculous mycobacterial infections - a comparative analysis of epidemiology, diagnosis and treatment.J Biomed Sci. 2020 Jun 17;27(1):74. doi: 10.1186/s12929-020-00667-6. J Biomed Sci. 2020. PMID: 32552732 Free PMC article. Review.
-
Mycobacterial Pathogenomics and Evolution.Microbiol Spectr. 2014 Feb;2(1):MGM2-0025-2013. doi: 10.1128/microbiolspec.MGM2-0025-2013. Microbiol Spectr. 2014. PMID: 26082120 Review.
Cited by
-
Mycobacteria that cause tuberculosis have retained ancestrally acquired genes for the biosynthesis of chemically diverse terpene nucleosides.PLoS Biol. 2024 Sep 30;22(9):e3002813. doi: 10.1371/journal.pbio.3002813. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39348416 Free PMC article.
References
-
- World Health Organization . 2023. Global tuberculosis report 2023. Geneva
-
- Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PDR, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, et al. . 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18:729–741. doi:10.1101/gr.075069.107 - DOI - PMC - PubMed
-
- Tortoli E, Fedrizzi T, Meehan CJ, Trovato A, Grottola A, Giacobazzi E, Serpini GF, Tagliazucchi S, Fabio A, Bettua C, Bertorelli R, Frascaro F, De Sanctis V, Pecorari M, Jousson O, Segata N, Cirillo DM. 2017. The new phylogeny of the genus Mycobacterium: the old and the news. Infect Genet Evol 56:19–25. doi:10.1016/j.meegid.2017.10.013 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

