Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infection
- PMID: 38701094
- PMCID: PMC11095722
- DOI: 10.1371/journal.ppat.1012205
Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infection
Abstract
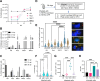
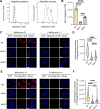
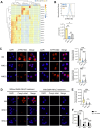
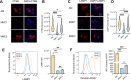
Mycobacterium tuberculosis (Mtb) infects lung myeloid cells, but the specific Mtb-permissive cells and host mechanisms supporting Mtb persistence during chronic infection are incompletely characterized. We report that after the development of T cell responses, CD11clo monocyte-derived cells harbor more live Mtb than alveolar macrophages (AM), neutrophils, and CD11chi monocyte-derived cells. Transcriptomic and functional studies revealed that the lysosome pathway is underexpressed in this highly permissive subset, characterized by less lysosome content, acidification, and proteolytic activity than AM, along with less nuclear TFEB, a regulator of lysosome biogenesis. Mtb infection does not drive lysosome deficiency in CD11clo monocyte-derived cells but promotes recruitment of monocytes that develop into permissive lung cells, mediated by the Mtb ESX-1 secretion system. The c-Abl tyrosine kinase inhibitor nilotinib activates TFEB and enhances lysosome functions of macrophages in vitro and in vivo, improving control of Mtb infection. Our results suggest that Mtb exploits lysosome-poor lung cells for persistence and targeting lysosome biogenesis is a potential host-directed therapy for tuberculosis.
Copyright: © 2024 Zheng et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Update of
-
Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infection.bioRxiv [Preprint]. 2023 May 28:2023.01.19.524758. doi: 10.1101/2023.01.19.524758. bioRxiv. 2023. Update in: PLoS Pathog. 2024 May 3;20(5):e1012205. doi: 10.1371/journal.ppat.1012205. PMID: 36711606 Free PMC article. Updated. Preprint.
-
Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infection.Res Sq [Preprint]. 2023 Jun 15:rs.3.rs-3049913. doi: 10.21203/rs.3.rs-3049913/v1. Res Sq. 2023. Update in: PLoS Pathog. 2024 May 3;20(5):e1012205. doi: 10.1371/journal.ppat.1012205. PMID: 37398178 Free PMC article. Updated. Preprint.
References
-
- Cambier CJ O’Leary SM, O’Sullivan MP, Keane J, Ramakrishnan L. Phenolic Glycolipid Facilitates Mycobacterial Escape from Microbicidal Tissue-Resident Macrophages. Immunity. 2017;47(3):552–65.e4. Epub 2017/08/29. doi: 10.1016/j.immuni.2017.08.003 ; PubMed Central PMCID: PMC5610147. - DOI - PMC - PubMed
-
- Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, et al. Alveolar Macrophages Provide an Early Mycobacterium tuberculosis Niche and Initiate Dissemination. Cell Host Microbe. 2018;24(3):439–46.e4. Epub 2018/08/28. doi: 10.1016/j.chom.2018.08.001 ; PubMed Central PMCID: PMC6152889. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

