scEWE: high-order element-wise weighted ensemble clustering for heterogeneity analysis of single-cell RNA-sequencing data
- PMID: 38701413
- PMCID: PMC11066953
- DOI: 10.1093/bib/bbae203
scEWE: high-order element-wise weighted ensemble clustering for heterogeneity analysis of single-cell RNA-sequencing data
Abstract
With the emergence of large amount of single-cell RNA sequencing (scRNA-seq) data, the exploration of computational methods has become critical in revealing biological mechanisms. Clustering is a representative for deciphering cellular heterogeneity embedded in scRNA-seq data. However, due to the diversity of datasets, none of the existing single-cell clustering methods shows overwhelming performance on all datasets. Weighted ensemble methods are proposed to integrate multiple results to improve heterogeneity analysis performance. These methods are usually weighted by considering the reliability of the base clustering results, ignoring the performance difference of the same base clustering on different cells. In this paper, we propose a high-order element-wise weighting strategy based self-representative ensemble learning framework: scEWE. By assigning different base clustering weights to individual cells, we construct and optimize the consensus matrix in a careful and exquisite way. In addition, we extracted the high-order information between cells, which enhanced the ability to represent the similarity relationship between cells. scEWE is experimentally shown to significantly outperform the state-of-the-art methods, which strongly demonstrates the effectiveness of the method and supports the potential applications in complex single-cell data analytical problems.
Keywords: Element-wise; Ensemble Clustering; High-order Similarity; scRNA-seq data.
© The Author(s) 2024. Published by Oxford University Press.
Figures
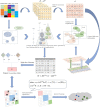
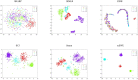



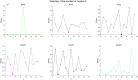


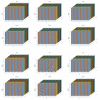

References
-
- Petegrosso R, Li Z, Kuang R. Machine learning and statistical methods for clustering single-cell rna-sequencing data. Brief Bioinform 2020;21(4):1209–23. - PubMed
-
- Hao J, Sohn LL, Haiyan H, Luonan C. Single cell clustering based on cell-pair differentiability correlation and variance analysis. Bioinformatics (Oxford, England) 34(21):3684–94. - PubMed
-
- Jiang H, Yi M, Zhang S. A kernel non-negative matrix factorization framework for single cell clustering. App Math Model 2021;90(1):875–88.

