The anti-leprosy drug clofazimine reduces polyQ toxicity through activation of PPARγ
- PMID: 38701619
- PMCID: PMC11088276
- DOI: 10.1016/j.ebiom.2024.105124
The anti-leprosy drug clofazimine reduces polyQ toxicity through activation of PPARγ
Abstract
Background: PolyQ diseases are autosomal dominant neurodegenerative disorders caused by the expansion of CAG repeats. While of slow progression, these diseases are ultimately fatal and lack effective therapies.
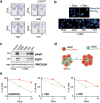
Methods: A high-throughput chemical screen was conducted to identify drugs that lower the toxicity of a protein containing the first exon of Huntington's disease (HD) protein huntingtin (HTT) harbouring 94 glutamines (Htt-Q94). Candidate drugs were tested in a wide range of in vitro and in vivo models of polyQ toxicity.
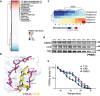
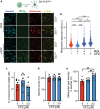
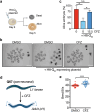
Findings: The chemical screen identified the anti-leprosy drug clofazimine as a hit, which was subsequently validated in several in vitro models. Computational analyses of transcriptional signatures revealed that the effect of clofazimine was due to the stimulation of mitochondrial biogenesis by peroxisome proliferator-activated receptor gamma (PPARγ). In agreement with this, clofazimine rescued mitochondrial dysfunction triggered by Htt-Q94 expression. Importantly, clofazimine also limited polyQ toxicity in developing zebrafish and neuron-specific worm models of polyQ disease.
Interpretation: Our results support the potential of repurposing the antimicrobial drug clofazimine for the treatment of polyQ diseases.
Funding: A full list of funding sources can be found in the acknowledgments section.
Keywords: Chemical screening; Huntington's disease; Mitochondria; PPARγ; polyQ.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Rawlins M.D., Wexler N.S., Wexler A.R., et al. The prevalence of Huntington's disease. Neuroepidemiology. 2016;46(2):144–153. - PubMed
-
- Kremer B., Goldberg P., Andrew S.E., et al. A worldwide study of the Huntington's disease mutation. The sensitivity and specificity of measuring CAG repeats. N Engl J Med. 1994;330(20):1401–1406. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

