H1FOO-DD promotes efficiency and uniformity in reprogramming to naive pluripotency
- PMID: 38701780
- PMCID: PMC11103934
- DOI: 10.1016/j.stemcr.2024.04.005
H1FOO-DD promotes efficiency and uniformity in reprogramming to naive pluripotency
Abstract
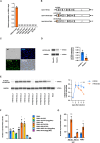
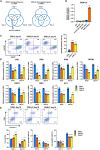
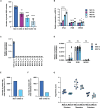
Heterogeneity among both primed and naive pluripotent stem cell lines remains a major unresolved problem. Here we show that expressing the maternal-specific linker histone H1FOO fused to a destabilizing domain (H1FOO-DD), together with OCT4, SOX2, KLF4, and LMYC, in human somatic cells improves the quality of reprogramming to both primed and naive pluripotency. H1FOO-DD expression was associated with altered chromatin accessibility around pluripotency genes and with suppression of the innate immune response. Notably, H1FOO-DD generates naive induced pluripotent stem cells with lower variation in transcriptome and methylome among clones and a more uniform and superior differentiation potency. Furthermore, we elucidated that upregulation of FKBP1A, driven by these five factors, plays a key role in H1FOO-DD-mediated reprogramming.
Keywords: FKBP1A; H1FOO; Sendai virus vector; destabilized domain; heterogeneity; induced pluripotent stem cell; innate immune response; naive pluripotency; primed pluripotency; reprogramming.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.K. and K.F. are co-inventors on a patent describing the method for producing human iPSCs from somatic cells using H1FOO-DD. K.F. is a co-founder and CEO of Heartseed Inc., and S.T., S. Yuasa, and K.F. own equity in Heartseed Inc. S.T. is an advisor of Heartseed Inc. J.K. and H.H. are employees and K.K. is a board member of ID Pharma Co., Ltd., without compensation relating to this study. S. Yamanaka is a scientific advisor to iPS Academia Japan without salary.
Figures



References
-
- Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science (New York, NY) 2004;303:1529–1531. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

