Molecular targets of glucocorticoids that elucidate their therapeutic efficacy in aggressive lymphomas
- PMID: 38701792
- PMCID: PMC11168741
- DOI: 10.1016/j.ccell.2024.04.007
Molecular targets of glucocorticoids that elucidate their therapeutic efficacy in aggressive lymphomas
Abstract
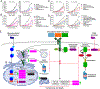
Glucocorticoids have been used for decades to treat lymphomas without an established mechanism of action. Using functional genomic, proteomic, and chemical screens, we discover that glucocorticoids inhibit oncogenic signaling by the B cell receptor (BCR), a recurrent feature of aggressive B cell malignancies, including diffuse large B cell lymphoma and Burkitt lymphoma. Glucocorticoids induce the glucocorticoid receptor (GR) to directly transactivate genes encoding negative regulators of BCR stability (LAPTM5; KLHL14) and the PI3 kinase pathway (INPP5D; DDIT4). GR directly represses transcription of CSK, a kinase that limits the activity of BCR-proximal Src-family kinases. CSK inhibition attenuates the constitutive BCR signaling of lymphomas by hyperactivating Src-family kinases, triggering their ubiquitination and degradation. With the knowledge that glucocorticoids disable oncogenic BCR signaling, they can now be deployed rationally to treat BCR-dependent aggressive lymphomas and used to construct mechanistically sound combination regimens with inhibitors of BTK, PI3 kinase, BCL2, and CSK.
Keywords: ABC; B cell receptor; CSK; DLBCL; GCB; PI3 kinase; combination therapy; dexamethasone; glucocorticoids; prednisone.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
-
- Rosenthal MC, Saunders RH, Schwartz LI, Zannos L, Perez Santiago E, and Dameshek W (1951). The use of adrenocorticotropic hormone and cortisone in the treatment of leukemia and leukosarcoma. Blood 6, 804–823. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

