Selection of a promiscuous minimalist cAMP phosphodiesterase from a library of de novo designed proteins
- PMID: 38702405
- PMCID: PMC11230910
- DOI: 10.1038/s41557-024-01490-4
Selection of a promiscuous minimalist cAMP phosphodiesterase from a library of de novo designed proteins
Abstract
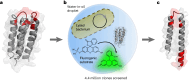
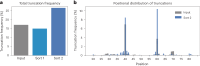
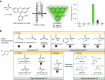
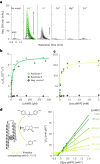
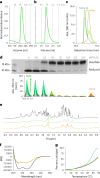
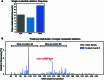
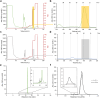
The ability of unevolved amino acid sequences to become biological catalysts was key to the emergence of life on Earth. However, billions of years of evolution separate complex modern enzymes from their simpler early ancestors. To probe how unevolved sequences can develop new functions, we use ultrahigh-throughput droplet microfluidics to screen for phosphoesterase activity amidst a library of more than one million sequences based on a de novo designed 4-helix bundle. Characterization of hits revealed that acquisition of function involved a large jump in sequence space enriching for truncations that removed >40% of the protein chain. Biophysical characterization of a catalytically active truncated protein revealed that it dimerizes into an α-helical structure, with the gain of function accompanied by increased structural dynamics. The identified phosphodiesterase is a manganese-dependent metalloenzyme that hydrolyses a range of phosphodiesters. It is most active towards cyclic AMP, with a rate acceleration of ~109 and a catalytic proficiency of >1014 M-1, comparable to larger enzymes shaped by billions of years of evolution.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
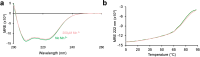

Similar articles
-
UCR1 and UCR2 domains unique to the cAMP-specific phosphodiesterase family form a discrete module via electrostatic interactions.J Biol Chem. 2000 Apr 7;275(14):10349-58. doi: 10.1074/jbc.275.14.10349. J Biol Chem. 2000. PMID: 10744723
-
Characterization of a novel cAMP-binding, cAMP-specific cyclic nucleotide phosphodiesterase (TcrPDEB1) from Trypanosoma cruzi.Biochem J. 2006 Oct 15;399(2):305-14. doi: 10.1042/BJ20060757. Biochem J. 2006. PMID: 16776650 Free PMC article.
-
DdPDE4, a novel cAMP-specific phosphodiesterase at the surface of dictyostelium cells.J Biol Chem. 2006 Jul 21;281(29):20018-26. doi: 10.1074/jbc.M600040200. Epub 2006 Apr 27. J Biol Chem. 2006. PMID: 16644729
-
[Phosphodiesterases of cyclic GMP].Postepy Hig Med Dosw. 2001;55(5):611-27. Postepy Hig Med Dosw. 2001. PMID: 11795198 Review. Polish.
-
cAMP-Specific phosphodiesterase-4 enzymes in the cardiovascular system: a molecular toolbox for generating compartmentalized cAMP signaling.Circ Res. 2007 Apr 13;100(7):950-66. doi: 10.1161/01.RES.0000261934.56938.38. Circ Res. 2007. PMID: 17431197 Review.
Cited by
-
Emergence of specific binding and catalysis from a designed generalist binding protein.bioRxiv [Preprint]. 2025 Jul 22:2025.01.30.635804. doi: 10.1101/2025.01.30.635804. bioRxiv. 2025. PMID: 39975260 Free PMC article. Preprint.
-
Droplet microfluidic screening to engineer angiotensin-converting enzyme 2 (ACE2) catalytic activity.J Biol Eng. 2025 Feb 3;19(1):12. doi: 10.1186/s13036-025-00482-3. J Biol Eng. 2025. PMID: 39901286 Free PMC article.
-
Fluorogenic, Subsingle-Turnover Monitoring of Enzymatic Reactions Involving NAD(P)H Provides a Generalized Platform for Directed Ultrahigh-Throughput Evolution of Biocatalysts in Microdroplets.J Am Chem Soc. 2025 Apr 2;147(13):10903-10915. doi: 10.1021/jacs.4c11804. Epub 2025 Mar 24. J Am Chem Soc. 2025. PMID: 40127491 Free PMC article.
-
From specialization to broad adoption: Key trends in droplet microfluidic innovations enhancing accessibility to non-experts.Biomicrofluidics. 2025 Mar 3;19(2):021302. doi: 10.1063/5.0242599. eCollection 2025 Mar. Biomicrofluidics. 2025. PMID: 40046719 Free PMC article.
References
-
- Tiessen A, Pérez-Rodríguez P, Delaye-Arredondo LJ. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res. Notes. 2012;5:85. - PMC - PubMed
-
- Mandecki W. A method for construction of long randomized open reading frames and polypeptides. Protein Eng. 1990;3:221–226. - PubMed
-
- Prijambada ID, et al. Solubility of artificial proteins with random sequences. FEBS Lett. 1996;382:21–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 98182/Volkswagen Foundation (VolkswagenStiftung)
- 695669/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- BB/W000504/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- MCB-1947720/National Science Foundation (NSF)
- P5R5PB_210999/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
LinkOut - more resources
Full Text Sources

