Viable protoplast isolation, organelle visualization and transformation of the globally distributed plant pathogen Phytophthora cinnamomi
- PMID: 38702562
- PMCID: PMC11358197
- DOI: 10.1007/s00709-024-01953-y
Viable protoplast isolation, organelle visualization and transformation of the globally distributed plant pathogen Phytophthora cinnamomi
Abstract
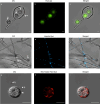
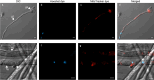
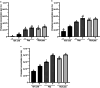
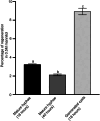
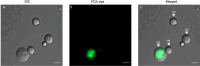
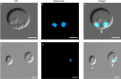
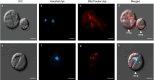
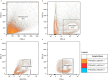
Phytophthora cinnamomi is an oomycete plant pathogen with a host range of almost 5000 plant species worldwide and therefore poses a serious threat to biodiversity. Omics technology has provided significant progress in our understanding of oomycete biology, however, transformation studies of Phytophthora for gene functionalisation are still in their infancy. Only a limited number of Phytophthora species have been successfully transformed and gene edited to elucidate the role of particular genes. There is a need to escalate our efforts to understand molecular processes, gene regulation and infection mechanisms of the pathogen to enable us to develop new disease management strategies. The primary obstacle hindering the advancement of transformation studies in Phytophthora is their challenging and unique nature, coupled with our limited comprehension of why they remain such an intractable system to work with. In this study, we have identified some of the key factors associated with the recalcitrant nature of P. cinnamomi. We have incorporated fluorescence microscopy and flow cytometry along with the organelle-specific dyes, fluorescein diacetate, Hoechst 33342 and MitoTracker™ Red CMXRos, to assess P. cinnamomi-derived protoplast populations. This approach has also provided valuable insights into the broader cell biology of Phytophthora. Furthermore, we have optimized the crucial steps that allow transformation of P. cinnamomi and have generated transformed isolates that express a cyan fluorescent protein, with a transformation efficiency of 19.5%. We therefore provide a platform for these methodologies to be applied for the transformation of other Phytophthora species and pave the way for future gene functionalisation studies.
Keywords: Phytophthora cinnamomi; Flow cytometry; Fluorescence microscopy; PEG/CaCl2 transformation; Protoplast.
© 2024. The Author(s).
Conflict of interest statement
The authors have no competing interests to declare that are relevant to the content of this article.
Figures


Similar articles
-
Transient expression of fluorescent proteins and Cas nucleases in Phytophthora agathidicida via PEG-mediated protoplast transformation.Microbiology (Reading). 2025 Mar;171(3):001547. doi: 10.1099/mic.0.001547. Microbiology (Reading). 2025. PMID: 40153308 Free PMC article.
-
An Improved Transformation System for Phytophthora cinnamomi Using Green Fluorescent Protein.Front Microbiol. 2021 Jul 5;12:682754. doi: 10.3389/fmicb.2021.682754. eCollection 2021. Front Microbiol. 2021. PMID: 34290684 Free PMC article.
-
PcAvh87, a virulence essential RxLR effector of Phytophthora cinnamomi suppresses host defense and induces cell death in plant nucleus.Microbiol Res. 2024 Sep;286:127789. doi: 10.1016/j.micres.2024.127789. Epub 2024 Jun 10. Microbiol Res. 2024. PMID: 38870619
-
In silico characterization of molecular factors involved in metabolism and pathogenicity of Phytophthora cinnamomi.Mol Biol Rep. 2022 Feb;49(2):1463-1473. doi: 10.1007/s11033-021-06901-0. Epub 2021 Nov 9. Mol Biol Rep. 2022. PMID: 34751913 Review.
-
Phytopathogenic oomycetes: a review focusing on Phytophthora cinnamomi and biotechnological approaches.Mol Biol Rep. 2020 Nov;47(11):9179-9188. doi: 10.1007/s11033-020-05911-8. Epub 2020 Oct 17. Mol Biol Rep. 2020. PMID: 33068230 Review.
Cited by
-
Transient expression of fluorescent proteins and Cas nucleases in Phytophthora agathidicida via PEG-mediated protoplast transformation.Microbiology (Reading). 2025 Mar;171(3):001547. doi: 10.1099/mic.0.001547. Microbiology (Reading). 2025. PMID: 40153308 Free PMC article.
-
Advancements in plant transformation: from traditional methods to cutting-edge techniques and emerging model species.Plant Cell Rep. 2024 Oct 29;43(11):273. doi: 10.1007/s00299-024-03359-9. Plant Cell Rep. 2024. PMID: 39467894 Review.
-
Viral vector-based transient expression systems for plant biotechnology research at PUIs.Front Educ (Lausanne). 2025;10:1598673. doi: 10.3389/feduc.2025.1598673. Epub 2025 Jun 1. Front Educ (Lausanne). 2025. PMID: 40520985 Free PMC article.
References
-
- Allardyce JA, Rookes JE, Cahill DM (2012) Defining plant resistance to Phytophthora cinnamomi: a standardized approach to assessment. J Phytopathol 160(6):269–276. 10.1111/j.1439-0434.2012.01895.x 10.1111/j.1439-0434.2012.01895.x - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

