LST-AI: A deep learning ensemble for accurate MS lesion segmentation
- PMID: 38703470
- PMCID: PMC11088188
- DOI: 10.1016/j.nicl.2024.103611
LST-AI: A deep learning ensemble for accurate MS lesion segmentation
Abstract
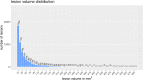
Automated segmentation of brain white matter lesions is crucial for both clinical assessment and scientific research in multiple sclerosis (MS). Over a decade ago, we introduced an engineered lesion segmentation tool, LST. While recent lesion segmentation approaches have leveraged artificial intelligence (AI), they often remain proprietary and difficult to adopt. As an open-source tool, we present LST-AI, an advanced deep learning-based extension of LST that consists of an ensemble of three 3D U-Nets. LST-AI explicitly addresses the imbalance between white matter (WM) lesions and non-lesioned WM. It employs a composite loss function incorporating binary cross-entropy and Tversky loss to improve segmentation of the highly heterogeneous MS lesions. We train the network ensemble on 491 MS pairs of T1-weighted and FLAIR images, collected in-house from a 3T MRI scanner, and expert neuroradiologists manually segmented the utilized lesion maps for training. LST-AI also includes a lesion location annotation tool, labeling lesions as periventricular, infratentorial, and juxtacortical according to the 2017 McDonald criteria, and, additionally, as subcortical. We conduct evaluations on 103 test cases consisting of publicly available data using the Anima segmentation validation tools and compare LST-AI with several publicly available lesion segmentation models. Our empirical analysis shows that LST-AI achieves superior performance compared to existing methods. Its Dice and F1 scores exceeded 0.62, outperforming LST, SAMSEG (Sequence Adaptive Multimodal SEGmentation), and the popular nnUNet framework, which all scored below 0.56. Notably, LST-AI demonstrated exceptional performance on the MSSEG-1 challenge dataset, an international WM lesion segmentation challenge, with a Dice score of 0.65 and an F1 score of 0.63-surpassing all other competing models at the time of the challenge. With increasing lesion volume, the lesion detection rate rapidly increased with a detection rate of >75% for lesions with a volume between 10 mm3 and 100 mm3. Given its higher segmentation performance, we recommend that research groups currently using LST transition to LST-AI. To facilitate broad adoption, we are releasing LST-AI as an open-source model, available as a command-line tool, dockerized container, or Python script, enabling diverse applications across multiple platforms.
Keywords: Artificial Intelligence; Deep Learning; Lesion Segmentation; Magnetic Resonance Imaging; Multiple Sclerosis; White Matter Lesions.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




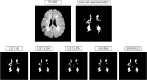


Update of
-
LST-AI: a Deep Learning Ensemble for Accurate MS Lesion Segmentation.medRxiv [Preprint]. 2024 Mar 11:2023.11.23.23298966. doi: 10.1101/2023.11.23.23298966. medRxiv. 2024. Update in: Neuroimage Clin. 2024;42:103611. doi: 10.1016/j.nicl.2024.103611. PMID: 38045345 Free PMC article. Updated. Preprint.
References
-
- Antonelli M., Reinke A., Bakas S., Farahani K., Kopp-Schneider A., Landman B.A., Litjens G., Menze B., Ronneberger O., Summers R.M., van Ginneken B., Bilello M., Bilic P., Christ P.F., Do R.K.G., Gollub M.J., Heckers S.H., Huisman H., Jarnagin W.R., Cardoso M.J. The medical segmentation decathlon. NatureCommunications. 2022;13(1):4128. doi: 10.1038/s41467-022-30695-9. - DOI - PMC - PubMed
-
- Ashtari P., Barile B., Van Huffel S., Sappey-Marinier D. New multiple sclerosis lesion segmentation and detection using pre-activation U-Net. Front. Neurosci. 2022;16 https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins... - DOI - PMC - PubMed
-
- Billot, B., Cerri, S., Leemput, K. V., Dalca, A. V., & Iglesias, J. E. (2021). Joint Segmentation Of Multiple Sclerosis Lesions And Brain Anatomy In MRI Scans Of Any Contrast And Resolution With CNNs. 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), 1971–1974. https://doi.org/10.1109/ISBI48211.2021.9434127. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

