Genetic variants for head size share genes and pathways with cancer
- PMID: 38703765
- PMCID: PMC11148644
- DOI: 10.1016/j.xcrm.2024.101529
Genetic variants for head size share genes and pathways with cancer
Abstract
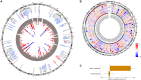
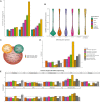
The size of the human head is highly heritable, but genetic drivers of its variation within the general population remain unmapped. We perform a genome-wide association study on head size (N = 80,890) and identify 67 genetic loci, of which 50 are novel. Neuroimaging studies show that 17 variants affect specific brain areas, but most have widespread effects. Gene set enrichment is observed for various cancers and the p53, Wnt, and ErbB signaling pathways. Genes harboring lead variants are enriched for macrocephaly syndrome genes (37-fold) and high-fidelity cancer genes (9-fold), which is not seen for human height variants. Head size variants are also near genes preferentially expressed in intermediate progenitor cells, neural cells linked to evolutionary brain expansion. Our results indicate that genes regulating early brain and cranial growth incline to neoplasia later in life, irrespective of height. This warrants investigation of clinical implications of the link between head size and cancer.
Keywords: cancer; genetics; genome-wide association study; head circumference; head size; intracranial volume; meta-analysis.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests H.H. and I.C.H. received funding from Alzheimer’s Research UK and the Dunhill Medical Trust Foundation. M.A.P. reported receiving grants and personal and travel fees from Roche, Novartis, Merck, and Biogen outside the submitted work. M. Scholz receives funding from Pfizer Inc. for a project not related to this research. C.D. serves as a consultant of Novartis Pharmaceuticals. B.F. has received educational speaking fees from Medice. N.J. and P.M.T. are MPIs of a research grant from Biogen Inc. for work unrelated to the contents of this manuscript. D.J.W. received funding from the Stroke Foundation/British Heart Foundation. D.J.S. has received consultancy honoraria from Discovery Vitality, Johnson & Johnson, Kanna, L’Oreal, Lundbeck, Orion, Sanofi, Servier, Takeda, and Vistagen. H.H. received funding from MRC, Wellcome Trust, and NIHR UCLH BRC. H.J.G. has received travel grants and speaker’s honoraria from Fresenius Medical Care, Neuraxpharm, and Janssen Cilag as well as research funding from Fresenius Medical Care.
Figures



References
-
- Dekaban A.S. Changes in brain weights during the span of human life: relation of brain weights to body heights and body weights. Ann. Neurol. 1978;4:345–356. - PubMed
-
- Smit D.J.A., Luciano M., Bartels M., van Beijsterveldt C.E.M., Wright M.J., Hansell N.K., Brunner H.G., Estourgie-van Burk G.F., de Geus E.J.C., Martin N.G., Boomsma D.I. Heritability of head size in Dutch and Australian twin families at ages 0-50 years. Twin Res. Hum. Genet. 2010;13:370–380. - PubMed
MeSH terms
Grants and funding
- P20 GM103472/GM/NIGMS NIH HHS/United States
- R01 NS017950/NS/NINDS NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- R01 DA033369/DA/NIDA NIH HHS/United States
- R01 AG058921/AG/NIA NIH HHS/United States
- R01 NS058700/NS/NINDS NIH HHS/United States
- F32 HL085989/HL/NHLBI NIH HHS/United States
- R01 DA031579/DA/NIDA NIH HHS/United States
- R01 HL067348/HL/NHLBI NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- U01 AG058589/AG/NIA NIH HHS/United States
- IK6 CX002519/CX/CSRD VA/United States
- 75N92023D00002/HL/NHLBI NIH HHS/United States
- 75N92023D00003/HL/NHLBI NIH HHS/United States
- R01 HL092301/HL/NHLBI NIH HHS/United States
- R01 AG049789/AG/NIA NIH HHS/United States
- R01 AG054076/AG/NIA NIH HHS/United States
- 75N92023D00005/HL/NHLBI NIH HHS/United States
- R01 AG016495/AG/NIA NIH HHS/United States
- RF1 AG059421/AG/NIA NIH HHS/United States
- R01 NS087541/NS/NINDS NIH HHS/United States
- R01 MH094524/MH/NIMH NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- 75N92023D00004/HL/NHLBI NIH HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- R01 AG049607/AG/NIA NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- R01 AG033040/AG/NIA NIH HHS/United States
- M01 RR007122/RR/NCRR NIH HHS/United States
- R01 NS075107/NS/NINDS NIH HHS/United States
- U24 RR021992/RR/NCRR NIH HHS/United States
- U01 AG052409/AG/NIA NIH HHS/United States
- 75N92019D00031/HL/NHLBI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- P30 AG010129/AG/NIA NIH HHS/United States
- U24 RR025736/RR/NCRR NIH HHS/United States
- P30 AG072972/AG/NIA NIH HHS/United States
- R01 HL087641/HL/NHLBI NIH HHS/United States
- 75N92023D00006/HL/NHLBI NIH HHS/United States
- U01 AG049505/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

