Targeting low levels of MIF expression as a potential therapeutic strategy for ALS
- PMID: 38703766
- PMCID: PMC11148722
- DOI: 10.1016/j.xcrm.2024.101546
Targeting low levels of MIF expression as a potential therapeutic strategy for ALS
Abstract
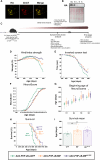
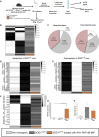
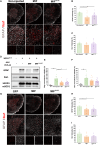
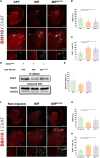
Mutations in SOD1 cause amyotrophic lateral sclerosis (ALS), a neurodegenerative disease characterized by motor neuron (MN) loss. We previously discovered that macrophage migration inhibitory factor (MIF), whose levels are extremely low in spinal MNs, inhibits mutant SOD1 misfolding and toxicity. In this study, we show that a single peripheral injection of adeno-associated virus (AAV) delivering MIF into adult SOD1G37R mice significantly improves their motor function, delays disease progression, and extends survival. Moreover, MIF treatment reduces neuroinflammation and misfolded SOD1 accumulation, rescues MNs, and corrects dysregulated pathways as observed by proteomics and transcriptomics. Furthermore, we reveal low MIF levels in human induced pluripotent stem cell-derived MNs from familial ALS patients with different genetic mutations, as well as in post mortem tissues of sporadic ALS patients. Our findings indicate that peripheral MIF administration may provide a potential therapeutic mechanism for modulating misfolded SOD1 in vivo and disease outcome in ALS patients.
Keywords: AAV; ALS; MIF; familial ALS; iPSCs; misfolded SOD1; motor neurons; mutant SOD1; mutant SOD1 mouse; sporadic ALS.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Cleveland D.W., Rothstein J.D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2001;2:806–819. - PubMed
-
- Abu-Hamad S., Kahn J., Leyton-Jaimes M.F., Rosenblatt J., Israelson A. Misfolded SOD1 Accumulation and Mitochondrial Association Contribute to the Selective Vulnerability of Motor Neurons in Familial ALS: Correlation to Human Disease. ACS Chem. Neurosci. 2017;8:2225–2234. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

