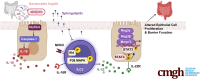
Bacterial Sphingolipids Exacerbate Colitis by Inhibiting ILC3-derived IL-22 Production
- PMID: 38704148
- PMCID: PMC11222953
- DOI: 10.1016/j.jcmgh.2024.04.007
Bacterial Sphingolipids Exacerbate Colitis by Inhibiting ILC3-derived IL-22 Production
Abstract
Background & aims: Gut bacterial sphingolipids, primarily produced by Bacteroidetes, have dual roles as bacterial virulence factors and regulators of the host mucosal immune system, including regulatory T cells and invariant natural killer T cells. Patients with inflammatory bowel disease display altered sphingolipids profiles in fecal samples. However, how bacterial sphingolipids modulate mucosal homeostasis and regulate intestinal inflammation remains unclear.
Methods: We used dextran sodium sulfate (DSS)-induced colitis in mice monocolonized with Bacteroides fragilis strains expressing or lacking sphingolipids to assess the influence of bacterial sphingolipids on intestinal inflammation using transcriptional, protein, and cellular analyses. Colonic explant and organoid were used to study the function of bacterial sphingolipids. Host mucosal immune cells and cytokines were profiled and characterized using flow cytometry, enzyme-linked immunosorbent assay, and Western blot, and cytokine function in vivo was investigated by monoclonal antibody injection.
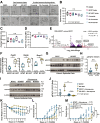
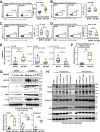
Results: B fragilis sphingolipids exacerbated intestinal inflammation. Mice monocolonized with B fragilis lacking sphingolipids exhibited less severe DSS-induced colitis. This amelioration of colitis was associated with increased production of interleukin (IL)-22 by ILC3. Mice colonized with B fragilis lacking sphingolipids following DSS treatment showed enhanced epithelial STAT3 activity, intestinal cell proliferation, and antimicrobial peptide production. Protection against DSS colitis associated with B fragilis lacking sphingolipids was reversed on IL22 blockade. Furthermore, bacterial sphingolipids restricted epithelial IL18 production following DSS treatment and interfered with IL22 production by a subset of ILC3 cells expressing both IL18R and major histocompatibility complex class II.
Conclusions: B fragilis-derived sphingolipids exacerbate mucosal inflammation by impeding epithelial IL18 expression and concomitantly suppressing the production of IL22 by ILC3 cells.
Keywords: Colitis; IL22; ILC3 Cells; Microbiota; Sphingolipids.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

