A Dataset for Evaluating Contextualized Representation of Biomedical Concepts in Language Models
- PMID: 38704422
- PMCID: PMC11069517
- DOI: 10.1038/s41597-024-03317-w
A Dataset for Evaluating Contextualized Representation of Biomedical Concepts in Language Models
Abstract
Due to the complexity of the biomedical domain, the ability to capture semantically meaningful representations of terms in context is a long-standing challenge. Despite important progress in the past years, no evaluation benchmark has been developed to evaluate how well language models represent biomedical concepts according to their corresponding context. Inspired by the Word-in-Context (WiC) benchmark, in which word sense disambiguation is reformulated as a binary classification task, we propose a novel dataset, BioWiC, to evaluate the ability of language models to encode biomedical terms in context. BioWiC comprises 20'156 instances, covering over 7'400 unique biomedical terms, making it the largest WiC dataset in the biomedical domain. We evaluate BioWiC both intrinsically and extrinsically and show that it could be used as a reliable benchmark for evaluating context-dependent embeddings in biomedical corpora. In addition, we conduct several experiments using a variety of discriminative and generative large language models to establish robust baselines that can serve as a foundation for future research.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

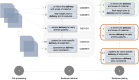

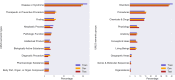

References
-
- Detroja, K., Bhensdadia, C. & Bhatt, B. S. A survey on relation extraction. Intell. Syst. with Appl. 200244 (2023).
-
- Shi, J. et al. Knowledge-graph-enabled biomedical entity linking: a survey. World Wide Web 1–30 (2023).
-
- Yazdani, A., Proios, D., Rouhizadeh, H. & Teodoro, D. Efficient joint learning for clinical named entity recognition and relation extraction using Fourier networks:a use case in adverse drug events. In Akhtar, M. S. & Chakraborty, T. (eds.) Proceedings of the 19th International Conference on Natural Language Processing (ICON), 212–223 (Association for Computational Linguistics, New Delhi, India, 2022)
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

