NRN1 interacts with Notch to increase oncogenic STAT3 signaling in melanoma
- PMID: 38705997
- PMCID: PMC11071257
- DOI: 10.1186/s12964-024-01632-8
NRN1 interacts with Notch to increase oncogenic STAT3 signaling in melanoma
Abstract
Background: Melanoma is a highly heterogeneous cancer, in which frequent changes in activation of signaling pathways lead to a high adaptability to ever changing tumor microenvironments. The elucidation of cancer specific signaling pathways is of great importance, as demonstrated by the inhibitor of the common BrafV600E mutation PLX4032 in melanoma treatment. We therefore investigated signaling pathways that were influenced by neurotrophin NRN1, which has been shown to be upregulated in melanoma.
Methods: Using a cell culture model system with an NRN1 overexpression, we investigated the influence of NRN1 on melanoma cells' functionality and signaling. We employed real time cell analysis and spheroid formation assays, while for investigation of molecular mechanisms we used a kinase phosphorylation kit as well as promotor activity analysis followed by mRNA and protein analysis.
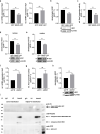
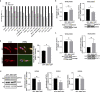
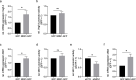
Results: We revealed that NRN1 interacts directly with the cleaved intracellular domain (NICD) of Notch1 and Notch3, causing a potential retention of NICD in the cytoplasm and thereby reducing the expression of its direct downstream target Hes1. This leads to decreased sequestration of JAK and STAT3 in a Hes1-driven phosphorylation complex. Consequently, our data shows less phosphorylation of STAT3 while presenting an accumulation of total protein levels of STAT3 in association with NRN1 overexpression. The potential of the STAT3 signaling pathway to act in both a tumor suppressive and oncogenic manner led us to investigate specific downstream targets - namely Vegf A, Mdr1, cMet - which were found to be upregulated under oncogenic levels of NRN1.
Conclusions: In summary, we were able to show that NRN1 links oncogenic signaling events between Notch and STAT3 in melanoma. We also suggest that in future research more attention should be payed to cellular regulation of signaling molecules outside of the classically known phosphorylation events.
Keywords: Melanoma; NRN1; Notch; STAT3.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

