Transcriptional profiles associated with coronary artery disease in type 2 diabetes mellitus
- PMID: 38706700
- PMCID: PMC11066158
- DOI: 10.3389/fendo.2024.1323168
Transcriptional profiles associated with coronary artery disease in type 2 diabetes mellitus
Abstract
Background: Coronary artery disease (CAD) is a common complication of Type 2 diabetes mellitus (T2DM). Understanding the pathogenesis of this complication is essential in both diagnosis and management. Thus, this study aimed to characterize the presence of CAD in T2DM using molecular markers and pathway analyses.
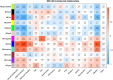
Methods: The study is a sex- and age-frequency matched case-control design comparing 23 unrelated adult Filipinos with T2DM-CAD to 23 controls (DM with CAD). Healthy controls served as a reference. Total RNA from peripheral blood mononuclear cells (PBMCs) underwent whole transcriptomic profiling using the Illumina HumanHT-12 v4.0 expression beadchip. Differential gene expression with gene ontogeny analyses was performed, with supporting correlational analyses using weighted correlation network analysis (WGCNA).
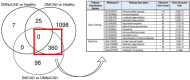
Results: The study observed that 458 genes were differentially expressed between T2DM with and without CAD (FDR<0.05). The 5 top genes the transcription factor 3 (TCF3), allograft inflammatory factor 1 (AIF1), nuclear factor, interleukin 3 regulated (NFIL3), paired immunoglobulin-like type 2 receptor alpha (PILRA), and cytoskeleton-associated protein 4 (CKAP4) with AUCs >89%. Pathway analyses show differences in innate immunity activity, which centers on the myelocytic (neutrophilic/monocytic) theme. SNP-module analyses point to a possible causal dysfunction in innate immunity that triggers the CAD injury in T2DM.
Conclusion: The study findings indicate the involvement of innate immunity in the development of T2DM-CAD, and potential immunity markers can reflect the occurrence of this injury. Further studies can verify the mechanistic hypothesis and use of the markers.
Keywords: coronary artery disease; differential gene expression; pathway analysis; transcriptomics; type 2 diabetes mellitus.
Copyright © 2024 Nevado, Cutiongco-de la Paz, Paz-Pacheco, Jasul, Aman, Deguit, San Pedro and Francisco.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

