Glucose starvation causes ferroptosis-mediated lysosomal dysfunction
- PMID: 38706843
- PMCID: PMC11067335
- DOI: 10.1016/j.isci.2024.109735
Glucose starvation causes ferroptosis-mediated lysosomal dysfunction
Abstract
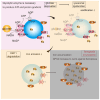
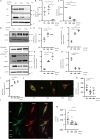
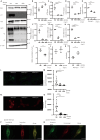
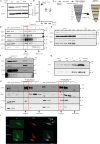
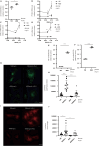
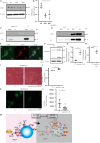
Lysosomes, the hub of metabolic signaling, are associated with various diseases and participate in autophagy by supplying nutrients to cells under nutrient starvation. However, their function and regulation under glucose starvation remain unclear and are studied herein. Under glucose starvation, lysosomal protein expression decreased, leading to the accumulation of damaged lysosomes. Subsequently, cell death occurred via ferroptosis and iron accumulation due to DMT1 degradation. GPX4, a key factor in ferroptosis inhibition located on the outer membrane of lysosomes, accumulated in lysosomes, especially under glucose starvation, to protect cells from ferroptosis. ALDOA, GAPDH, NAMPT, and PGK1 are also located on the outer membrane of lysosomes and participate in lysosomal function. These enzymes did not function effectively under glucose starvation, leading to lysosomal dysfunction and ferroptosis. These findings may facilitate the treatment of lysosomal-related diseases.
Keywords: Cell biology; Cellular physiology; Functional aspects of cell biology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

