Uncovering specific taxonomic and functional alteration of gut microbiota in chronic kidney disease through 16S rRNA data
- PMID: 38707511
- PMCID: PMC11066246
- DOI: 10.3389/fcimb.2024.1363276
Uncovering specific taxonomic and functional alteration of gut microbiota in chronic kidney disease through 16S rRNA data
Abstract
Introduction: Chronic kidney disease (CKD) is worldwide healthcare burden with growing incidence and death rate. Emerging evidence demonstrated the compositional and functional differences of gut microbiota in patients with CKD. As such, gut microbial features can be developed as diagnostic biomarkers and potential therapeutic target for CKD.
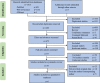
Methods: To eliminate the outcome bias arising from factors such as geographical distribution, sequencing platform, and data analysis techniques, we conducted a comprehensive analysis of the microbial differences between patients with CKD and healthy individuals based on multiple samples worldwide. A total of 980 samples from six references across three nations were incorporated from the PubMed, Web of Science, and GMrepo databases. The obtained 16S rRNA microbiome data were subjected to DADA2 processing, QIIME2 and PICRUSt2 analyses.
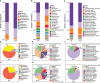
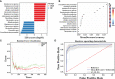
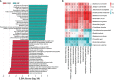
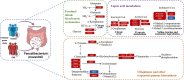
Results: The gut microbiota of patients with CKD differs significantly from that of healthy controls (HC), with a substantial decrease in the microbial diversity among the CKD group. Moreover, a significantly reduced abundance of bacteria Faecalibacterium prausnitzii (F. prausnitzii) was detected in the CKD group through linear discriminant analysis effect size (LEfSe) analysis, which may be associated with the alleviating effects against CKD. Notably, we identified CKD-depleted F. prausnitzii demonstrated a significant negative correlation with three pathways based on predictive functional analysis, suggesting its potential role in regulating systemic acidbase disturbance and pro-oxidant metabolism.
Discussion: Our findings demonstrated notable alterations of gut microbiota in CKD patients. Specific gut-beneficial microbiota, especially F. prausnitzii, may be developed as a preventive and therapeutic tool for CKD clinical management.
Keywords: 16S rRNA; biomarker; chronic kidney disease; gut microbiota; probiotics.
Copyright © 2024 Zhang, Zhong, Liu, Wang, Lin, Lin, Fang, Mou, Jiang, Huang, Zhao and Zheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Alterations to the Gut Microbiota and Their Correlation With Inflammatory Factors in Chronic Kidney Disease.Front Cell Infect Microbiol. 2019 Jun 12;9:206. doi: 10.3389/fcimb.2019.00206. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31245306 Free PMC article.
-
Adolescent gut microbiome imbalance and its association with immune response in inflammatory bowel diseases and obesity.BMC Microbiol. 2024 Jul 19;24(1):268. doi: 10.1186/s12866-024-03425-y. BMC Microbiol. 2024. PMID: 39030520 Free PMC article.
-
Characterizing the gut microbiota in patients with chronic kidney disease.Postgrad Med. 2020 Aug;132(6):495-505. doi: 10.1080/00325481.2020.1744335. Epub 2020 Apr 2. Postgrad Med. 2020. PMID: 32241215
-
Specific alterations in gut microbiota in patients with chronic kidney disease: an updated systematic review.Ren Fail. 2021 Dec;43(1):102-112. doi: 10.1080/0886022X.2020.1864404. Ren Fail. 2021. PMID: 33406960 Free PMC article.
-
Gut microbiota alterations in colorectal adenoma-carcinoma sequence based on 16S rRNA gene sequencing: A systematic review and meta-analysis.Microb Pathog. 2024 Oct;195:106889. doi: 10.1016/j.micpath.2024.106889. Epub 2024 Aug 26. Microb Pathog. 2024. PMID: 39197689
Cited by
-
The evolving understanding of systemic mechanisms in organ-specific IgA nephropathy: a focus on gut-kidney crosstalk.Theranostics. 2025 Jan 1;15(2):656-681. doi: 10.7150/thno.104631. eCollection 2025. Theranostics. 2025. PMID: 39744688 Free PMC article. Review.
-
Evaluation of the Correlation between Gut Microbiota and Renal Function in Chronic Kidney Disease Patients.J Microbiol Biotechnol. 2025 Jun 17;35:e2502039. doi: 10.4014/jmb.2502.02039. J Microbiol Biotechnol. 2025. PMID: 40537901 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

