Network and Experimental Pharmacology on Mechanism of Yixintai Regulates the TMAO/PKC/NF-κB Signaling Pathway in Treating Heart Failure
- PMID: 38707614
- PMCID: PMC11069381
- DOI: 10.2147/DDDT.S448140
Network and Experimental Pharmacology on Mechanism of Yixintai Regulates the TMAO/PKC/NF-κB Signaling Pathway in Treating Heart Failure
Abstract
Objective: This study aims to explore the mechanism of action of Yixintai in treating chronic ischemic heart failure by combining bioinformatics and experimental validation.
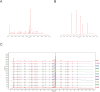
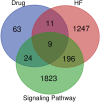
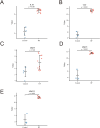
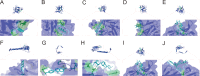
Materials and methods: Five potential drugs for treating heart failure were obtained from Yixintai (YXT) through early mass spectrometry detection. The targets of YXT for treating heart failure were obtained by a search of online databases. Gene ontology (GO) functional enrichment analysis and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses were conducted on the common targets using the DAVID database. A rat heart failure model was established by ligating the anterior descending branch of the left coronary artery. A small animal color Doppler ultrasound imaging system detected cardiac function indicators. Hematoxylin-eosin (HE), Masson's, and electron microscopy were used to observe the pathological morphology of the myocardium in rats with heart failure. The network pharmacology analysis results were validated by ELISA, qPCR, and Western blotting.
Results: A total of 107 effective targets were obtained by combining compound targets and eliminating duplicate values. PPI analysis showed that inflammation-related proteins (TNF and IL1B) were key targets for treating heart failure, and KEGG enrichment suggested that NF-κB signaling pathway was a key pathway for YXT treatment of heart failure. Animal model validation results indicated the following: YXT can significantly reduce the content of intestinal microbiota metabolites such as trimethylamine oxide (TMAO) and improve heart failure by improving the EF and FS values of heart ultrasound in rats and reducing the levels of serum NT-proBNP, ANP, and BNP to improve heart failure. Together, YXT can inhibit cardiac muscle hypertrophy and fibrosis in rats and improve myocardial ultrastructure and serum IL-1β, IL-6, and TNF-α levels. These effects are achieved by inhibiting the expressions of NF-κB and PKC.
Conclusion: YXT regulates the TMAO/PKC/NF-κB signaling pathway in heart failure.
Keywords: PKC/NF-κB pathway; TMAO; heart failure; network pharmacology; traditional Chinese medicine; yixintai.
© 2024 Wang et al.
Conflict of interest statement
The authors declare no competing interests in this work.
Figures









References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

