Testing of putative antiseizure medications in a preclinical Dravet syndrome zebrafish model
- PMID: 38707709
- PMCID: PMC11069116
- DOI: 10.1093/braincomms/fcae135
Testing of putative antiseizure medications in a preclinical Dravet syndrome zebrafish model
Abstract
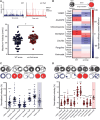
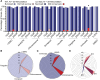
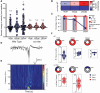
Dravet syndrome is a severe genetic epilepsy primarily caused by de novo mutations in a voltage-activated sodium channel gene (SCN1A). Patients face life-threatening seizures that are largely resistant to available anti-seizure medications. Preclinical Dravet syndrome animal models are a valuable tool to identify candidate anti-seizure medications for these patients. Among these, scn1lab mutant zebrafish, exhibiting spontaneous seizure-like activity, are particularly amenable to large-scale drug screening. Thus far, we have screened more than 3000 drug candidates in scn1lab zebrafish mutants, identifying valproate, stiripentol, and fenfluramine e.g. Food and Drug Administration-approved drugs, with clinical application in the Dravet syndrome population. Successful phenotypic screening in scn1lab mutant zebrafish is rigorous and consists of two stages: (i) a locomotion-based assay measuring high-velocity convulsive swim behaviour and (ii) an electrophysiology-based assay, using in vivo local field potential recordings, to quantify electrographic seizure-like events. Historically, nearly 90% of drug candidates fail during translation from preclinical models to the clinic. With such a high failure rate, it becomes necessary to address issues of replication and false positive identification. Leveraging our scn1lab zebrafish assays is one approach to address these problems. Here, we curated a list of nine anti-seizure drug candidates recently identified by other groups using preclinical Dravet syndrome models: 1-Ethyl-2-benzimidazolinone, AA43279, chlorzoxazone, donepezil, lisuride, mifepristone, pargyline, soticlestat and vorinostat. First-stage locomotion-based assays in scn1lab mutant zebrafish identified only 1-Ethyl-2-benzimidazolinone, chlorzoxazone and lisuride. However, second-stage local field potential recording assays did not show significant suppression of spontaneous electrographic seizure activity for any of the nine anti-seizure drug candidates. Surprisingly, soticlestat induced frank electrographic seizure-like discharges in wild-type control zebrafish. Taken together, our results failed to replicate clear anti-seizure efficacy for these drug candidates highlighting a necessity for strict scientific standards in preclinical identification of anti-seizure medications.
Keywords: anti-seizure medications; dravet; drug screening; epilepsy; zebrafish larvae.
Published by Oxford University Press on behalf of the Guarantors of Brain 2024.
Conflict of interest statement
The authors declare the following competing interests: S.C.B is a co-Founder and Chief Scientific Advisor for Epygenix Therapeutics. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Update of
-
Testing of putative antiseizure drugs in a preclinical Dravet syndrome zebrafish model.bioRxiv [Preprint]. 2023 Nov 15:2023.11.11.566723. doi: 10.1101/2023.11.11.566723. bioRxiv. 2023. Update in: Brain Commun. 2024 Apr 16;6(3):fcae135. doi: 10.1093/braincomms/fcae135. PMID: 38014342 Free PMC article. Updated. Preprint.
References
-
- Marini C, Scheffer IE, Nabbout R, et al. SCN1A duplications and deletions detected in Dravet syndrome: Implications for molecular diagnosis. Epilepsia. 2009;50(7):1670–1678. - PubMed
-
- Mei D, Cetica V, Marini C, Guerrini R. Dravet syndrome as part of the clinical and genetic spectrum of sodium channel epilepsies and encephalopathies. Epilepsia. 2019;60(Suppl 3):S2–S7. - PubMed
-
- Dravet C. Dravet syndrome history. Dev Med Child Neurol. 2011;53(Suppl 2):1–6. - PubMed
-
- Scheffer IE, Nabbout R. SCN1A-related phenotypes: Epilepsy and beyond. Epilepsia. 2019;60(Suppl 3):S17–S24. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
