Correlation between Mitochondria-Associated Endoplasmic Reticulum Membrane-Related Genes and Cellular Senescence-Related Genes in Osteoarthritis
- PMID: 38708239
- PMCID: PMC11064197
- DOI: 10.1021/acsomega.3c10316
Correlation between Mitochondria-Associated Endoplasmic Reticulum Membrane-Related Genes and Cellular Senescence-Related Genes in Osteoarthritis
Abstract
Background: The role of mitochondria-associated endoplasmic reticulum membrane (MAM) formation in the development of osteoarthritis (OA) is yet unclear.
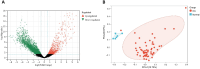
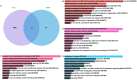
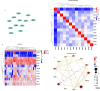
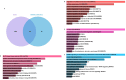
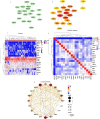
Methods: A mix of bioinformatics methods and in vitro experimental methodologies was used to study and corroborate the role of MAM-related genes and cellular senescence-related genes in the development of OA. The Gene Expression Omnibus database was used to obtain the microarray information that is relevant to the OA. Several bioinformatic methods were employed to carry out function enrichment analysis and protein-protein correlation analysis, build the correlation regulatory network, and investigate potential relationships between MAM-related genes and cellular senescence-related genes in OA. These methods also served to identify the MAM-related and OA-related genes (MAM-OARGs).
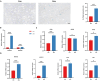
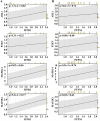
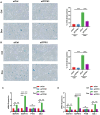
Results: For the additional functional enrichment analysis, a total of 13 MAM-OARGs were detected. The correlation regulatory network was also created. Hub MAM-OARGs were shown to have a strong correlation with genes relevant to cellular senescence in OA. Results of in vitro experiments further demonstrated a positive correlation between MAM-OARGs (PTPN1 and ITPR1) and cellular senescence-related and OA-related genes.
Conclusions: As a result, our findings can offer new insights into the investigations of MAM-related genes and cellular senescence-related genes, which could be linked to the OA as well as brand-new potential treatment targets.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Zhao B.; Zhang Q.; He Y.; Cao W.; Song W.; Liang X. Targeted metabolomics reveals the aberrant energy status in diabetic peripheral neuropathy and the neuroprotective mechanism of traditional Chinese medicine JinMaiTong. J. Pharm. Anal. 2024, 14 (2), 225–243. 10.1016/j.jpha.2023.09.007. - DOI - PMC - PubMed
-
- Liang X.-L.; Ouyang L.; Yu N.-N.; Sun Z.-H.; Gui Z.-K.; Niu Y.-L.; He Q.-Y.; Zhang J.; Wang Y. Histone deacetylase inhibitor pracinostat suppresses colorectal cancer by inducing CDK5-Drp1 signaling-mediated peripheral mitofission. J. Pharm. Anal. 2023, 13 (10), 1168–1182. 10.1016/j.jpha.2023.06.005. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
