Cost-effort analysis of Baited Remote Underwater Video (BRUV) and environmental DNA (eDNA) in monitoring marine ecological communities
- PMID: 38708339
- PMCID: PMC11067900
- DOI: 10.7717/peerj.17091
Cost-effort analysis of Baited Remote Underwater Video (BRUV) and environmental DNA (eDNA) in monitoring marine ecological communities
Abstract
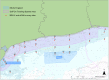
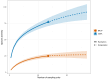
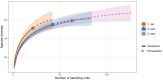
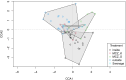
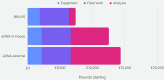
Monitoring the diversity and distribution of species in an ecosystem is essential to assess the success of restoration strategies. Implementing biomonitoring methods, which provide a comprehensive assessment of species diversity and mitigate biases in data collection, holds significant importance in biodiversity research. Additionally, ensuring that these methods are cost-efficient and require minimal effort is crucial for effective environmental monitoring. In this study we compare the efficiency of species detection, the cost and the effort of two non-destructive sampling techniques: Baited Remote Underwater Video (BRUV) and environmental DNA (eDNA) metabarcoding to survey marine vertebrate species. Comparisons were conducted along the Sussex coast upon the introduction of the Nearshore Trawling Byelaw. This Byelaw aims to boost the recovery of the dense kelp beds and the associated biodiversity that existed in the 1980s. We show that overall BRUV surveys are more affordable than eDNA, however, eDNA detects almost three times as many species as BRUV. eDNA and BRUV surveys are comparable in terms of effort required for each method, unless eDNA analysis is carried out externally, in which case eDNA requires less effort for the lead researchers. Furthermore, we show that increased eDNA replication yields more informative results on community structure. We found that using both methods in conjunction provides a more complete view of biodiversity, with BRUV data supplementing eDNA monitoring by recording species missed by eDNA and by providing additional environmental and life history metrics. The results from this study will serve as a baseline of the marine vertebrate community in Sussex Bay allowing future biodiversity monitoring research projects to understand community structure as the ecosystem recovers following the removal of trawling fishing pressure. Although this study was regional, the findings presented herein have relevance to marine biodiversity and conservation monitoring programs around the globe.
Keywords: Baseline survey; Biodiversity monitoring; Coastal ecosystem; Marine biodiversity; Population ecology.
©2024 Clark et al.
Conflict of interest statement
Nathan R. Geraldi works for NatureMetrics.
Figures


References
-
- Afzali SF, Bourdages H, Laporte M, Mérot C, Normandeau E, Audet C, Bernatchez L. Comparing environmental metabarcoding and trawling survey of demersal fish communities in the Gulf of St. Lawrence, Canada. Environmental DNA. 2021;3:22–42. doi: 10.1002/edn3.111. - DOI
-
- Alexander JB, Bunce M, White N, Wilkinson SP, Adam AAS, Berry T, Stat M, Thomas L, Newman SJ, Dugal L, Richards ZT. Development of a multi-assay approach for monitoring coral diversity using eDNA metabarcoding. Coral Reefs. 2020;39:159–171. doi: 10.1007/s00338-019-01875-9. - DOI
-
- Alfaro-Cordova E, Ortiz-Alvarez C, Alfaro-Shigueto J, Mangel JC, García O, Velez-Zuazo X. What lies beneath? Revealing biodiversity through eDNA analysis in Lobos de Afuera Islands, Peru. Latin American Journal of Aquatic Research. 2022;50:642–659. doi: 10.3856/vol50-issue4-fulltext-2782. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

