Multiple E3 ligases act as antiviral factors against SARS-CoV-2 via inducing the ubiquitination and degradation of ORF9b
- PMID: 38709105
- PMCID: PMC11237466
- DOI: 10.1128/jvi.01624-23
Multiple E3 ligases act as antiviral factors against SARS-CoV-2 via inducing the ubiquitination and degradation of ORF9b
Abstract
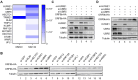
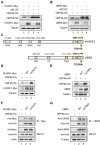
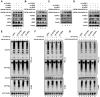
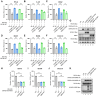
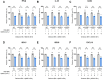
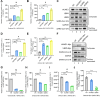
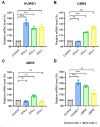
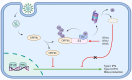
Severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) open reading frame 9b (ORF9b) antagonizes the antiviral type I and III interferon (IFN) responses and is ubiquitinated and degraded via the ubiquitin-proteasome pathway. However, E3 ubiquitin ligases that mediate the polyubiquitination and degradation of ORF9b remain unknown. In this study, we identified 14 E3 ligases that specifically bind to SARS-CoV-2 ORF9b. Specifically, three E3 ligases, HECT, UBA, and WWE domain-containing E3 ubiquitin protein ligase 1 (HUWE1), ubiquitin protein ligase E3 component n-recognin 4 (UBR4), and UBR5, induced K48-linked polyubiquitination and degradation of ORF9b, thereby attenuating ORF9b-mediated inhibition of the IFN response and SARS-CoV-2 replication. Moreover, each E3 ligase performed this function independent of the other two E3 ligases. Therefore, the three E3 ligases identified in this study as anti-SARS-CoV-2 host factors provide novel molecular insight into the virus-host interaction.IMPORTANCEUbiquitination is an important post-translational modification that regulates multiple biological processes, including viral replication. Identification of E3 ubiquitin ligases that target viral proteins for degradation can provide novel targets for antagonizing viral infections. Here, we identified multiple E3 ligases, including HECT, UBA, and WWE domain-containing E3 ubiquitin protein ligase 1 (HUWE1), ubiquitin protein ligase E3 component n-recognin 4 (UBR4), and UBR5, that ubiquitinated and induced the degradation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) open reading frame 9b (ORF9b), an interferon (IFN) antagonist, thereby enhancing IFN production and attenuating SARS-CoV-2 replication. Our study provides new possibilities for drug development targeting the interaction between E3 ligases and ORF9b.
Keywords: HUWE1; SARS-CoV-2 ORF9b; UBR4; UBR5; degradation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
UBR5 Acts as an Antiviral Host Factor against MERS-CoV via Promoting Ubiquitination and Degradation of ORF4b.J Virol. 2022 Sep 14;96(17):e0074122. doi: 10.1128/jvi.00741-22. Epub 2022 Aug 18. J Virol. 2022. PMID: 35980206 Free PMC article.
-
The Deubiquitinase USP29 Promotes SARS-CoV-2 Virulence by Preventing Proteasome Degradation of ORF9b.mBio. 2022 Jun 28;13(3):e0130022. doi: 10.1128/mbio.01300-22. Epub 2022 May 31. mBio. 2022. PMID: 35638730 Free PMC article.
-
SARS-CoV-2 nsp16 is regulated by host E3 ubiquitin ligases, UBR5 and MARCHF7.Elife. 2025 May 13;13:RP102277. doi: 10.7554/eLife.102277. Elife. 2025. PMID: 40358464 Free PMC article.
-
Human E3 ubiquitin ligases: accelerators and brakes for SARS-CoV-2 infection.Biochem Soc Trans. 2024 Oct 30;52(5):2009-2021. doi: 10.1042/BST20230324. Biochem Soc Trans. 2024. PMID: 39222407 Free PMC article. Review.
-
Spatial and temporal roles of SARS-CoV PLpro -A snapshot.FASEB J. 2021 Jan;35(1):e21197. doi: 10.1096/fj.202002271. FASEB J. 2021. PMID: 33368679 Free PMC article. Review.
Cited by
-
Extended insights into the pathophysiological role of UBR5: a commentary.Int J Surg. 2024 Sep 1;110(9):6024-6025. doi: 10.1097/JS9.0000000000001716. Int J Surg. 2024. PMID: 39275785 Free PMC article. No abstract available.
-
SARS-CoV-2 enhances complement-mediated endothelial injury via the suppression of membrane complement regulatory proteins.Emerg Microbes Infect. 2025 Dec;14(1):2467781. doi: 10.1080/22221751.2025.2467781. Epub 2025 Feb 28. Emerg Microbes Infect. 2025. PMID: 39945674 Free PMC article.
-
Genetic diversity and genomic epidemiology of SARS-CoV-2 during the first 3 years of the pandemic in Morocco: comprehensive sequence analysis, including the unique lineage B.1.528 in Morocco.Access Microbiol. 2024 Oct 7;6(10):000853.v4. doi: 10.1099/acmi.0.000853.v4. eCollection 2024. Access Microbiol. 2024. PMID: 39376591 Free PMC article.
-
SARS-CoV-2-ORF-3a Mediates Apoptosis Through Mitochondrial Dysfunction Modulated by the K+ Ion Channel.Int J Mol Sci. 2025 Feb 13;26(4):1575. doi: 10.3390/ijms26041575. Int J Mol Sci. 2025. PMID: 40004042 Free PMC article.
-
ZYG11B suppresses multiple enteroviruses by triggering viral VP1 degradation.J Virol. 2025 Apr 15;99(4):e0003025. doi: 10.1128/jvi.00030-25. Epub 2025 Mar 26. J Virol. 2025. PMID: 40135890 Free PMC article.
References
-
- Zhou Y, Zheng R, Liu S, Disoma C, Du A, Li S, Chen Z, Dong Z, Zhang Y, Li S, Liu P, Razzaq A, Chen X, Liao Y, Tao S, Liu Y, Xu L, Zhang Q, Peng J, Deng X, Li S, Jiang T, Xia Z. 2022. Host E3 Ligase HUWE1 attenuates the proapoptotic activity of the MERS-CoV accessory protein ORF3 by promoting its ubiquitin-dependent degradation. J Biol Chem 298:101584. doi:10.1016/j.jbc.2022.101584 - DOI - PMC - PubMed
-
- Wu J, Shi Y, Pan X, Wu S, Hou R, Zhang Y, Zhong T, Tang H, Du W, Wang L, Wo J, Mu J, Qiu Y, Yang K, Zhang LK, Ye BC, Qi N. 2021. SARS-CoV-2 ORF9B inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep 34:108761. doi:10.1016/j.celrep.2021.108761 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 81930062, 82272316/MOST | National Natural Science Foundation of China (NSFC)
- 2021YFC2301900, 2021YFC2301904, 20210101300JC/MOST | National Key Research and Development Program of China (NKPs)
- YDZJ202301ZYTS521/Department of Science and Technology of Jilin Province ()
- YDZJ202201ZYTS587/Department of Science and Technology of Jilin Province ()
- 20102209/Jilin Province Key Laboratory of Molecular Virology
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

