Cooltools: Enabling high-resolution Hi-C analysis in Python
- PMID: 38709825
- PMCID: PMC11098495
- DOI: 10.1371/journal.pcbi.1012067
Cooltools: Enabling high-resolution Hi-C analysis in Python
Abstract
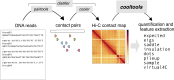
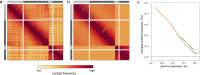
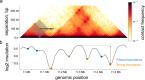
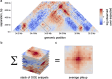
Chromosome conformation capture (3C) technologies reveal the incredible complexity of genome organization. Maps of increasing size, depth, and resolution are now used to probe genome architecture across cell states, types, and organisms. Larger datasets add challenges at each step of computational analysis, from storage and memory constraints to researchers' time; however, analysis tools that meet these increased resource demands have not kept pace. Furthermore, existing tools offer limited support for customizing analysis for specific use cases or new biology. Here we introduce cooltools (https://github.com/open2c/cooltools), a suite of computational tools that enables flexible, scalable, and reproducible analysis of high-resolution contact frequency data. Cooltools leverages the widely-adopted cooler format which handles storage and access for high-resolution datasets. Cooltools provides a paired command line interface (CLI) and Python application programming interface (API), which respectively facilitate workflows on high-performance computing clusters and in interactive analysis environments. In short, cooltools enables the effective use of the latest and largest genome folding datasets.
Copyright: © 2024 Open2C et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

