Metagenomics datasets of water and sediments from eutrophication-impacted artificial lakes in South Africa
- PMID: 38710672
- PMCID: PMC11074141
- DOI: 10.1038/s41597-024-03286-0
Metagenomics datasets of water and sediments from eutrophication-impacted artificial lakes in South Africa
Abstract
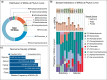
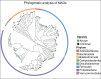
We present metagenomes of 16 samples of water and sediment from two lakes, collected from eutrophic and non-eutrophic areas, including pooled samples enriched with phosphate and nitrate. Additionally, we assembled 167 bacterial metagenome-assembled genomes (MAGs). These MAGs were de-replicated into 83 unique genomes representing different species found in the lakes. All the MAGs exhibited >70% completeness and <10% contamination, with 79 MAGs being classified as 'nearly complete' (completeness >90%), while 54 falling within 80-90% range and 34 between 75-80% complete. The most abundant MAGs identified across all samples were Proteobacteria (n = 80), Firmicutes_A (n = 35), Firmicutes (n = 13), and Bacteriodota (n = 22). Other groups included Desulfobacteria_I (n = 2), Verrucomicrobiota (n = 4), Campylobacterota (n = 4) and Actinobacteriota (n = 6). Importantly, phylogenomic analysis identified that approximately 50.3% of the MAGs could not be classified to known species, suggesting the presence of potentially new and unknown bacteria in these lakes, warranting further in-depth investigation. This study provides valuable new dataset on the diverse and often unique microbial communities living in polluted lakes, useful in developing effective strategies to manage pollution.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Stats South Africa. Interactive population distribution maps of the Republic of South Africa (2023). (2023).
-
- O’Brien GC, et al. River connectivity and fish migration considerations in the management of multiple stressors in South Africa. Mar. Freshw. Res. 2019;70:1254–1264. doi: 10.1071/MF19183. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

