Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms
- PMID: 38710674
- PMCID: PMC11074275
- DOI: 10.1038/s41467-024-48277-2
Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms
Abstract
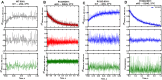
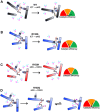
Mutations in human isocitrate dehydrogenase 1 (IDH1) drive tumor formation in a variety of cancers by replacing its conventional activity with a neomorphic activity that generates an oncometabolite. Little is understood of the mechanistic differences among tumor-driving IDH1 mutants. We previously reported that the R132Q mutant unusually preserves conventional activity while catalyzing robust oncometabolite production, allowing an opportunity to compare these reaction mechanisms within a single active site. Here, we employ static and dynamic structural methods and observe that, compared to R132H, the R132Q active site adopts a conformation primed for catalysis with optimized substrate binding and hydride transfer to drive improved conventional and neomorphic activity over R132H. This active site remodeling reveals a possible mechanism of resistance to selective mutant IDH1 therapeutic inhibitors. This work enhances our understanding of fundamental IDH1 mechanisms while pinpointing regions for improving inhibitor selectivity.
© 2024. The Author(s).
Conflict of interest statement
J.M.S. is an employee at Vividion Therapeutics and owns stock in Schrödinger. The remaining authors declare no competing interests.
Figures







Update of
-
Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.bioRxiv [Preprint]. 2024 Jan 23:2024.01.10.574970. doi: 10.1101/2024.01.10.574970. bioRxiv. 2024. Update in: Nat Commun. 2024 May 6;15(1):3785. doi: 10.1038/s41467-024-48277-2. PMID: 38260668 Free PMC article. Updated. Preprint.
-
Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.Res Sq [Preprint]. 2024 Feb 23:rs.3.rs-3889456. doi: 10.21203/rs.3.rs-3889456/v1. Res Sq. 2024. Update in: Nat Commun. 2024 May 6;15(1):3785. doi: 10.1038/s41467-024-48277-2. PMID: 38464189 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

