Pemigatinib in previously treated solid tumors with activating FGFR1-FGFR3 alterations: phase 2 FIGHT-207 basket trial
- PMID: 38710951
- PMCID: PMC11186762
- DOI: 10.1038/s41591-024-02934-7
Pemigatinib in previously treated solid tumors with activating FGFR1-FGFR3 alterations: phase 2 FIGHT-207 basket trial
Erratum in
-
Publisher Correction: Pemigatinib in previously treated solid tumors with activating FGFR1-FGFR3 alterations: phase 2 FIGHT-207 basket trial.Nat Med. 2024 Aug;30(8):2377. doi: 10.1038/s41591-024-03072-w. Nat Med. 2024. PMID: 38789647 Free PMC article. No abstract available.
Abstract
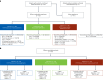
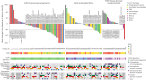
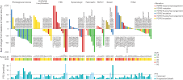
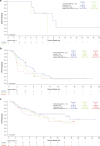
Fibroblast growth factor receptor (FGFR) alterations drive oncogenesis in multiple tumor types. Here we studied pemigatinib, a selective, potent, oral FGFR1-FGFR3 inhibitor, in the phase 2 FIGHT-207 basket study of FGFR-altered advanced solid tumors. Primary end points were objective response rate (ORR) in cohorts A (fusions/rearrangements, n = 49) and B (activating non-kinase domain mutations, n = 32). Secondary end points were progression-free survival, duration of response and overall survival in cohorts A and B, and safety. Exploratory end points included ORR of cohort C (kinase domain mutations, potentially pathogenic variants of unknown significance, n = 26) and analysis of co-alterations associated with resistance and response. ORRs for cohorts A, B and C were 26.5% (13/49), 9.4% (3/32) and 3.8% (1/26), respectively. Tumors with no approved FGFR inhibitors or those with alterations not previously confirmed to be sensitive to FGFR inhibition had objective responses. In cohorts A and B, the median progression-free survival was 4.5 and 3.7 months, median duration of response was 7.8 and 6.9 months and median overall survival was 17.5 and 11.4 months, respectively. Safety was consistent with previous reports. The most common any-grade treatment-emergent adverse events were hyperphosphatemia (84%) and stomatitis (53%). TP53 co-mutations were associated with lack of response and BAP1 alterations with higher response rates. FGFR1-FGFR3 gatekeeper and molecular brake mutations led to acquired resistance. New therapeutic areas for FGFR inhibition and drug failure mechanisms were identified across tumor types. ClinicalTrials.gov identifier: NCT03822117 .
© 2024. The Author(s).
Conflict of interest statement
J.R. served as a consultant or advisor for AADi, Avoro Capital Advisors, Boxer Capital, Chinese University of Hong Kong, Clarion Healthcare, Columbus Venture Partners, Cullgen, Debiopharm Group, Ellipses Pharma, Envision Pharma Group, Incyte, iOnctura, Macrogenics, Merus, Monte Rosa Therapeutics, Oncology One, Pfizer, Sardona Therapeutics, Vall d’Hebron Institute of Oncology/Ministerio de Empleo y Seguridad Social and Tang Advisors; received travel support from ESMO; received research funding paid directly to the institution from AADi, Amgen, Bayer, Bicycle Therapeutics, BioAtla, BioMed Valley Discoveries, Black Diamond Therapeutics, Blueprint Medicines, Cellestia Biotech, Curis, CytomX Therapeutics, Deciphera, Fore Biotherapeutics, Genmab, GlaxoSmithKline, Hummingbird, Hutchison MediPharma, IDEAYA Biosciences, Incyte, Kelun, Linnaeus Therapeutics, Loxo, Merck Sharp & Dohme, Merus, Mirati Therapeutics, Novartis, Nuvation Bio, Pfizer, Roche, Spectrum Pharmaceuticals, Symphogen, Taiho Pharmaceutical, Takeda/Millennium, Tango Therapeutics, Vall d’Hebron Institute of Oncology/Cancer Core Europe and Yingli Pharma; and reported a relationship with Vall d’Hebron Institute of Oncology/Ministerio de Empleo y Seguridad Social. S.D. received research funding paid directly to the institution from Basilea Pharmaceutica, Incyte, Nerviano Medical Science, Pfizer and Roche. M.F. received institutional research grants from AbbVie, Amgen, Aprea, AstraZeneca, Beigene, BMS, Checkmate, Elicio, Genmab, Gilead, GSK, Incyte, Jacobio, Lilly, Merck, Mirati and Novartis; served on advisory boards for AbbVie, AstraZeneca, Jazz Pharma, Beigene and Mirati; and is a consultant for Omega Therapeutics and Novartis. J.G.-D. received research funding from Astellas, BMS, GSK, Ipsen, Janssen, Pfizer, Roche and Sanofi and honoraria for serving as a speaker for AstraZeneca, BMS, Janssen and Roche. A.I. served on advisory boards for AstraZeneca, Bayer, Chugai, Daiichi Sankyo, GSK, Merck, MSD, Parthenon and Roche and received research grants from AstraZeneca, Bayer, BMS, GSK, Merck, MSD, Novartis, Parthenon, Pfizer and Roche. I.S. received institutional research grants from Alligator Bioscience, AstraZeneca, BMS, Cantargia AB, Genentech, Genmab, Incyte, Loxo/Bayer, Loxo/Lilly, MSD, Novartis, Orion, Roche, Pfizer, Puma Biotechnology and Symphogen and support for attending meetings and/or travel expenses from AstraZeneca, Incyte, Merck and Pfizer. M.U. received research grants from Astellas Pharma, AstraZeneca, Boehringer Ingelheim, CHUGAI Pharmaceutical, DFP, Eisai, Eli Lilly, Incyte, J-Pharma, Merck Biopharma, MSD, Novartis, Ono Pharmaceutical and Taiho Pharmaceutical and honoraria from AstraZeneca, CHUGAI Pharmaceutical, Eisai, Incyte, MSD, Novartis, Ono Pharmaceutical and Taiho Pharmaceutical. T.Y. received research grants from AbbVie, AMED, Ascent, AstraZeneca, GlaxoSmithKlineINBC, Incyte, Lilly, Merck Biopharma, MSD, Nanobiotix, Novartis, Ono Pharmaceutical, Pfizer, Roche and Syneos Health and lecture fees from AstraZeneca, Bristol-Meyers Squibb, Chugai, Eisai, Merck Biopharma, MSD, Ono Pharmaceutical and Rakuten Medical. M.L.V., N.O., X.L., A.G. and M.S. are employees and shareholders of Incyte. L.G. served on a data safety and monitoring committee for AstraZeneca and on advisory boards or as a consultant for Alentis Therapeutics AG, Black Diamond, Blueprint Medicine, Compass Therapeutics, Eisai/H3Biomedicine, Exelixis, Genentech, Kinnate, Incyte Corporation, Merck, QED Therapeutics, Servier, Sirtex Medical, Surface Oncology, Taiho Oncology, TranstheraBio, Tyra Biosciences, AbbVie, AstraZeneca and Cogent Biosciences.
Figures

References
-
- Incyte. PEMAZYRE (pemigatinib). Full prescribing information. (2022).
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

