Positive feedback regulation between glycolysis and histone lactylation drives oncogenesis in pancreatic ductal adenocarcinoma
- PMID: 38711083
- PMCID: PMC11071201
- DOI: 10.1186/s12943-024-02008-9
Positive feedback regulation between glycolysis and histone lactylation drives oncogenesis in pancreatic ductal adenocarcinoma
Abstract
Background: Metabolic reprogramming and epigenetic alterations contribute to the aggressiveness of pancreatic ductal adenocarcinoma (PDAC). Lactate-dependent histone modification is a new type of histone mark, which links glycolysis metabolite to the epigenetic process of lactylation. However, the role of histone lactylation in PDAC remains unclear.
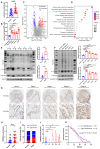
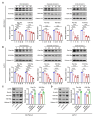
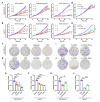
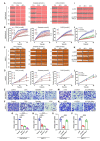
Methods: The level of histone lactylation in PDAC was identified by western blot and immunohistochemistry, and its relationship with the overall survival was evaluated using a Kaplan-Meier survival plot. The participation of histone lactylation in the growth and progression of PDAC was confirmed through inhibition of histone lactylation by glycolysis inhibitors or lactate dehydrogenase A (LDHA) knockdown both in vitro and in vivo. The potential writers and erasers of histone lactylation in PDAC were identified by western blot and functional experiments. The potential target genes of H3K18 lactylation (H3K18la) were screened by CUT&Tag and RNA-seq analyses. The candidate target genes TTK protein kinase (TTK) and BUB1 mitotic checkpoint serine/threonine kinase B (BUB1B) were validated through ChIP-qPCR, RT-qPCR and western blot analyses. Next, the effects of these two genes in PDAC were confirmed by knockdown or overexpression. The interaction between TTK and LDHA was identified by Co-IP assay.
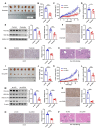
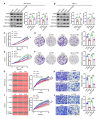
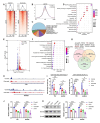
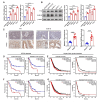
Results: Histone lactylation, especially H3K18la level was elevated in PDAC, and the high level of H3K18la was associated with poor prognosis. The suppression of glycolytic activity by different kinds of inhibitors or LDHA knockdown contributed to the anti-tumor effects of PDAC in vitro and in vivo. E1A binding protein p300 (P300) and histone deacetylase 2 were the potential writer and eraser of histone lactylation in PDAC cells, respectively. H3K18la was enriched at the promoters and activated the transcription of mitotic checkpoint regulators TTK and BUB1B. Interestingly, TTK and BUB1B could elevate the expression of P300 which in turn increased glycolysis. Moreover, TTK phosphorylated LDHA at tyrosine 239 (Y239) and activated LDHA, and subsequently upregulated lactate and H3K18la levels.
Conclusions: The glycolysis-H3K18la-TTK/BUB1B positive feedback loop exacerbates dysfunction in PDAC. These findings delivered a new exploration and significant inter-relationship between lactate metabolic reprogramming and epigenetic regulation, which might pave the way toward novel lactylation treatment strategies in PDAC therapy.
Keywords: Glycolysis; H3K18la; Histone lactylation; Pancreatic ductal adenocarcinoma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

