Identification and characterization of a new potent inhibitor targeting CtBP1/BARS in melanoma cells
- PMID: 38711119
- PMCID: PMC11071220
- DOI: 10.1186/s13046-024-03044-5
Identification and characterization of a new potent inhibitor targeting CtBP1/BARS in melanoma cells
Abstract
Background: The C-terminal-binding protein 1/brefeldin A ADP-ribosylation substrate (CtBP1/BARS) acts both as an oncogenic transcriptional co-repressor and as a fission inducing protein required for membrane trafficking and Golgi complex partitioning during mitosis, hence for mitotic entry. CtBP1/BARS overexpression, in multiple cancers, has pro-tumorigenic functions regulating gene networks associated with "cancer hallmarks" and malignant behavior including: increased cell survival, proliferation, migration/invasion, epithelial-mesenchymal transition (EMT). Structurally, CtBP1/BARS belongs to the hydroxyacid-dehydrogenase family and possesses a NAD(H)-binding Rossmann fold, which, depending on ligands bound, controls the oligomerization of CtBP1/BARS and, in turn, its cellular functions. Here, we proposed to target the CtBP1/BARS Rossmann fold with small molecules as selective inhibitors of mitotic entry and pro-tumoral transcriptional activities.
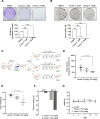
Methods: Structured-based screening of drug databases at different development stages was applied to discover novel ligands targeting the Rossmann fold. Among these identified ligands, N-(3,4-dichlorophenyl)-4-{[(4-nitrophenyl)carbamoyl]amino}benzenesulfonamide, called Comp.11, was selected for further analysis. Fluorescence spectroscopy, isothermal calorimetry, computational modelling and site-directed mutagenesis were employed to define the binding of Comp.11 to the Rossmann fold. Effects of Comp.11 on the oligomerization state, protein partners binding and pro-tumoral activities were evaluated by size-exclusion chromatography, pull-down, membrane transport and mitotic entry assays, Flow cytometry, quantitative real-time PCR, motility/invasion, and colony assays in A375MM and B16F10 melanoma cell lines. Effects of Comp.11 on tumor growth in vivo were analyzed in mouse tumor model.
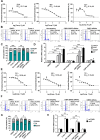
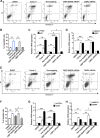
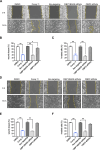
Results: We identify Comp.11 as a new, potent and selective inhibitor of CtBP1/BARS (but not CtBP2). Comp.11 directly binds to the CtBP1/BARS Rossmann fold affecting the oligomerization state of the protein (unlike other known CtBPs inhibitors), which, in turn, hinders interactions with relevant partners, resulting in the inhibition of both CtBP1/BARS cellular functions: i) membrane fission, with block of mitotic entry and cellular secretion; and ii) transcriptional pro-tumoral effects with significantly hampered proliferation, EMT, migration/invasion, and colony-forming capabilities. The combination of these effects impairs melanoma tumor growth in mouse models. CONCLUSIONS: This study identifies a potent and selective inhibitor of CtBP1/BARS active in cellular and melanoma animal models revealing new opportunities to study the role of CtBP1/BARS in tumor biology and to develop novel melanoma treatments.
Keywords: Benzenesulfonamide; C-terminal Binding Protein (CtBP); CtBP inhibitor; Melanoma; Rossmann fold.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

