This is a preprint.
Hair follicles modulate skin barrier function
- PMID: 38712094
- PMCID: PMC11071379
- DOI: 10.1101/2024.04.23.590728
Hair follicles modulate skin barrier function
Update in
-
Hair follicles modulate skin barrier function.Cell Rep. 2024 Jul 23;43(7):114347. doi: 10.1016/j.celrep.2024.114347. Epub 2024 Jun 26. Cell Rep. 2024. PMID: 38941190 Free PMC article.
Abstract
Our skin provides a protective barrier that shields us from our environment. Barrier function is typically associated with interfollicular epidermis; however, whether hair follicles influence this process remains unclear. Here, we utilize a potent genetic tool to probe barrier function by conditionally ablating a quintessential epidermal barrier gene, Abca12, which is mutated in the most severe skin barrier disease, harlequin ichthyosis. With this tool, we deduced 4 ways by which hair follicles modulate skin barrier function. First, the upper hair follicle (uHF) forms a functioning barrier. Second, barrier disruption in the uHF elicits non-cell autonomous responses in the epidermis. Third, deleting Abca12 in the uHF impairs desquamation and blocks sebum release. Finally, barrier perturbation causes uHF cells to move into the epidermis. Neutralizing Il17a, whose expression is enriched in the uHF, partially alleviated some disease phenotypes. Altogether, our findings implicate hair follicles as multi-faceted regulators of skin barrier function.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests. Artificial intelligence (A.I.) was not used for the preparation of this manuscript.
Figures
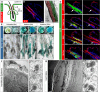
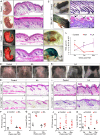
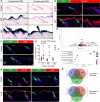

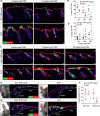


References
-
- Candi E., Schmidt R., and Melino G. (2005). The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6, 328–340. - PubMed
-
- Lippens S., Denecker G., Ovaere P., Vandenabeele P., and Declercq W. (2005). Death penalty for keratinocytes: apoptosis versus cornification. Cell Death Differ 12, 1497–1508. - PubMed
-
- Feingold K.R., and Elias P.M. (2014). Role of lipids in the formation and maintenance of the cutaneous permeability barrier. Biochim Biophys Acta 1841, 280–294. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
