This is a preprint.
CYpHER: Catalytic extracellular targeted protein degradation with high potency and durable effect
- PMID: 38712232
- PMCID: PMC11071310
- DOI: 10.1101/2024.02.21.581471
CYpHER: Catalytic extracellular targeted protein degradation with high potency and durable effect
Update in
-
CYpHER: catalytic extracellular targeted protein degradation with high potency and durable effect.Nat Commun. 2024 Oct 9;15(1):8731. doi: 10.1038/s41467-024-52975-2. Nat Commun. 2024. PMID: 39384759 Free PMC article.
Abstract
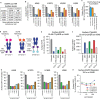
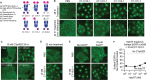
Many disease-causing proteins have multiple pathogenic mechanisms, and conventional inhibitors struggle to reliably disrupt more than one. Targeted protein degradation (TPD) can eliminate the protein, and thus all its functions, by directing a cell's protein turnover machinery towards it. Two established strategies either engage catalytic E3 ligases or drive uptake towards the endolysosomal pathway. Here we describe CYpHER (CatalYtic pH-dependent Endolysosomal delivery with Recycling) technology with potency and durability from a novel catalytic mechanism that shares the specificity and straightforward modular design of endolysosomal uptake. By bestowing pH-dependent release on the target engager and using the rapid-cycling transferrin receptor as the uptake receptor, CYpHER induces endolysosomal target delivery while re-using drug, potentially yielding increased potency and reduced off-target tissue exposure risks. The TfR-based approach allows targeting to tumors that overexpress this receptor and offers the potential for transport to the CNS. CYpHER function was demonstrated in vitro with EGFR and PD-L1, and in vivo with EGFR in a model of EGFR-driven non-small cell lung cancer.
Conflict of interest statement
Competing interests Cyclera Therapeutics Inc. retains intellectual property rights to the technology described in this manuscript. Z.R.C., G.P.S., and N.W.N. own stock in and are employees of Cyclera. J.M.O. owns stock in and is an advisor of Cyclera. Z.R.C., J.M.O., and N.W.N. are inventors on patent applications for this technology. P.Y., E.J.G., T-D.P., M.H., and J.P. have no competing interests.
Figures






References
-
- Chang M. T., Shanahan F., Nguyen T. T. T., Staben S. T., Gazzard L., Yamazoe S., Wertz I. E., Piskol R., Yang Y. A., Modrusan Z., Haley B., Evangelista M., Malek S., Foster S. A., Ye X., Identifying transcriptional programs underlying cancer drug response with TraCe-seq. Nat Biotechnol 40, 86–93 (2022). - PubMed
-
- Dai W., Li Y., Zhou Q., Xu Z., Sun C., Tan X., Lu L., Cetuximab inhibits oral squamous cell carcinoma invasion and metastasis via degradation of epidermal growth factor receptor. J Oral Pathology Medicine 43, 250–257 (2014). - PubMed
-
- Lu Y., Li X., Liang K., Luwor R., Siddik Z. H., Mills G. B., Mendelsohn J., Fan Z., Epidermal Growth Factor Receptor (EGFR) Ubiquitination as a Mechanism of Acquired Resistance Escaping Treatment by the Anti-EGFR Monoclonal Antibody Cetuximab. Cancer Research 67, 8240–8247 (2007). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
