CYRI-B-mediated macropinocytosis drives metastasis via lysophosphatidic acid receptor uptake
- PMID: 38712822
- PMCID: PMC11219039
- DOI: 10.7554/eLife.83712
CYRI-B-mediated macropinocytosis drives metastasis via lysophosphatidic acid receptor uptake
Abstract
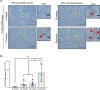
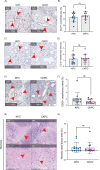
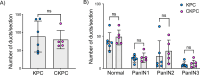
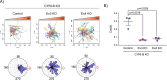
Pancreatic ductal adenocarcinoma carries a dismal prognosis, with high rates of metastasis and few treatment options. Hyperactivation of KRAS in almost all tumours drives RAC1 activation, conferring enhanced migratory and proliferative capacity as well as macropinocytosis. Macropinocytosis is well understood as a nutrient scavenging mechanism, but little is known about its functions in trafficking of signalling receptors. We find that CYRI-B is highly expressed in pancreatic tumours in a mouse model of KRAS and p53-driven pancreatic cancer. Deletion of Cyrib (the gene encoding CYRI-B protein) accelerates tumourigenesis, leading to enhanced ERK and JNK-induced proliferation in precancerous lesions, indicating a potential role as a buffer of RAC1 hyperactivation in early stages. However, as disease progresses, loss of CYRI-B inhibits metastasis. CYRI-B depleted tumour cells show reduced chemotactic responses to lysophosphatidic acid, a major driver of tumour spread, due to impaired macropinocytic uptake of the lysophosphatidic acid receptor 1. Overall, we implicate CYRI-B as a mediator of growth and signalling in pancreatic cancer, providing new insights into pathways controlling metastasis.
Keywords: actin cytoskeleton; cancer biology; cell biology; chemotaxis; endocytosis; macropinocytosis; metastasis; mouse; pancreatic cancer.
Plain language summary
Pancreatic cancer is an aggressive disease with limited treatment options. It is also associated with high rates of metastasis – meaning it spreads to other areas of the body. Environmental pressures, such as a lack of the nutrients metastatic cancer cells need to grow and divide, can change how the cells behave. Understanding the changes that allow cancer cells to respond to these pressures could reveal new treatment options for pancreatic cancer. When nutrients are scarce, metastatic cancer cells can gather molecules and nutrients by capturing large amounts of the fluid that surrounds them using a mechanism called macropinocytosis. They can also migrate to areas of the body with higher nutrient levels, through a process called chemotaxis. This involves cells moving towards areas with higher levels of certain molecules. For example, cancer cells migrate towards high levels of a lipid called lysophosphatidic acid, which promotes their growth and survival. A newly discovered protein known as CYRI-B has recently been shown to regulate how cells migrate and take up nutrients. It also interacts with proteins known to be involved in pancreatic cancer progression. Therefore, Nikolaou et al. set out to investigate whether CYRI-B also plays a role in metastatic pancreatic cancer. Experiments in a mouse model of pancreatic cancer showed that CYRI-B levels were high in pancreatic tumour cells. And when the gene for CYRI-B was removed from the tumour cells, they did not metastasise. Further analysis revealed that CYRI-B controls uptake and processing of nutrients and other signalling molecules through macropinocytosis. In particular, it ensures uptake of the receptor for lysophosphatidic acid, allowing the metastatic cancer cells to migrate. The findings of Nikolaou et al. reveal that CYRI-B is involved in metastasis of cancer cells in a mouse model of pancreatic cancer. This new insight into how metastasis is controlled could help to identify future targets for treatments that aim to prevent pancreatic cancer cells spreading to distant sites.
© 2024, Nikolaou et al.
Conflict of interest statement
SN, AJ, JW, NP, LF, CN, HS, SB, LM No competing interests declared
Figures





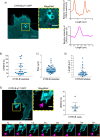




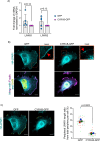

Update of
References
-
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJC, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UMH, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

