In vitro, in planta, and comparative genomic analyses of Pseudomonas syringae pv. syringae strains of pepper (Capsicum annuum var. annuum)
- PMID: 38712940
- PMCID: PMC11237606
- DOI: 10.1128/spectrum.00064-24
In vitro, in planta, and comparative genomic analyses of Pseudomonas syringae pv. syringae strains of pepper (Capsicum annuum var. annuum)
Abstract
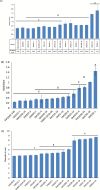
Pseudomonas syringae pv. syringae (Pss) is an emerging phytopathogen that causes Pseudomonas leaf spot (PLS) disease in pepper plants. Pss can cause serious economic damage to pepper production, yet very little is known about the virulence factors carried by Pss that cause disease in pepper seedlings. In this study, Pss strains isolated from pepper plants showing PLS symptoms in Ohio between 2013 and 2021 (n = 16) showed varying degrees of virulence (Pss populations and disease symptoms on leaves) on 6-week-old pepper seedlings. In vitro studies assessing growth in nutrient-limited conditions, biofilm production, and motility also showed varying degrees of virulence, but in vitro and in planta variation in virulence between Pss strains did not correlate. Comparative whole-genome sequencing studies identified notable virulence genes including 30 biofilm genes, 87 motility genes, and 106 secretion system genes. Additionally, a total of 27 antimicrobial resistance genes were found. A multivariate correlation analysis and Scoary analysis based on variation in gene content (n = 812 variable genes) and single nucleotide polymorphisms within virulence genes identified no significant correlations with disease severity, likely due to our limited sample size. In summary, our study explored the virulence and antimicrobial gene content of Pss in pepper seedlings as a first step toward understanding the virulence and pathogenicity of Pss in pepper seedlings. Further studies with additional pepper Pss strains will facilitate defining genes in Pss that correlate with its virulence in pepper seedlings, which can facilitate the development of effective measures to control Pss in pepper and other related P. syringae pathovars.
Importance: Pseudomonas leaf spot (PLS) caused by Pseudomonas syringae pv. syringae (Pss) causes significant losses to the pepper industry. Highly virulent Pss strains under optimal environmental conditions (cool-moderate temperatures, high moisture) can cause severe necrotic lesions on pepper leaves that consequently can decrease pepper yield if the disease persists. Hence, it is important to understand the virulence mechanisms of Pss to be able to effectively control PLS in peppers. In our study, in vitro, in planta, and whole-genome sequence analyses were conducted to better understand the virulence and pathogenicity characteristics of Pss strains in peppers. Our findings fill a knowledge gap regarding potential virulence and pathogenicity characteristics of Pss in peppers, including virulence and antimicrobial gene content. Our study helps pave a path to further identify the role of specific virulence genes in causing disease in peppers, which can have implications in developing strategies to effectively control PLS in peppers.
Keywords: Pseudomonas syringae pv. syringae; comparative genomic analysis; pepper plants; phytobacteria; virulence genes; whole-genome sequence analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Morris CE, Lamichhane JR, Nikolić I, Stanković S, Moury B. 2019. The overlapping continuum of host range among strains in the Pseudomonas syringae complex. Phytopathol Res 1:4. doi:10.1186/s42483-018-0010-6 - DOI
-
- Tripodi P, Kumar S. 2019. The capsicum crop: an introduction, p 1–8. In Ramchiary N, Kole C (ed), The capsicum genome. Springer International Publishing, Cham.
-
- Vegetables 2020 summary 02/11/2021 99. 2020.
-
- Lamichhane JR, Messéan A, Morris CE. 2015. Insights into epidemiology and control of diseases of annual plants caused by the Pseudomonas syringae species complex. J Gen Plant Pathol 81:331–350. doi:10.1007/s10327-015-0605-z - DOI
-
- Miller SA. 2019. Pepper diseases and their control. Great Lakes Expo.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

