Characterization of virus‒host recombinant variants of the hepatitis E virus
- PMID: 38712945
- PMCID: PMC11237545
- DOI: 10.1128/jvi.00295-24
Characterization of virus‒host recombinant variants of the hepatitis E virus
Abstract
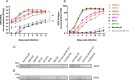
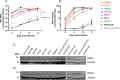
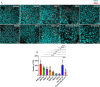
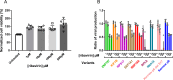
Hepatitis E virus is a single-strand, positive-sense RNA virus that can lead to chronic infection in immunocompromised patients. Virus-host recombinant variants (VHRVs) have been described in such patients. These variants integrate part of human genes into the polyproline-rich region that could introduce new post-translational modifications (PTMs), such as ubiquitination. The aim of this study was to characterize the replication capacity of different VHRVs, namely, RNF19A, ZNF787, KIF1B, EEF1A1, RNA18, RPS17, and RPL6. We used a plasmid encoding the Kernow strain, in which the fragment encoding the S17 insertion was deleted (Kernow p6 delS17) or replaced by fragments encoding the different insertions. The HEV RNA concentrations in the supernatants and the HepG2/C3A cell lysates were determined via RT-qPCR. The capsid protein ORF2 was immunostained. The effect of ribavirin was also assessed. The HEV RNA concentrations in the supernatants and the cell lysates were higher for the variants harboring the RNF19A, ZNF787, KIF1B, RPS17, and EEF1A1 insertions than for the Kernow p6 del S17, while it was not with RNA18 or RPL6 fragments. The number of ORF2 foci was higher for RNF19A, ZNF787, KIF1B, and RPS17 than for Kernow p6 del S17. VHRVs with replicative advantages were less sensitive to the antiviral effect of ribavirin. No difference in PTMs was found between VHRVs with a replicative advantage and those without. In conclusion, our study showed that insertions did not systematically confer a replicative advantage in vitro. Further studies are needed to determine the mechanisms underlying the differences in replicative capacity.
Importance: Hepatitis E virus (HEV) is a major cause of viral hepatitis. HEV can lead to chronic infection in immunocompromised patients. Ribavirin treatment is currently used to treat such chronic infections. Recently, seven virus-host recombinant viruses were characterized in immunocompromised patients. These viruses have incorporated a portion of a human gene fragment into their genome. We studied the consequences of these insertions on the replication capacity. We found that these inserted fragments could enhance virus replication for five of the seven recombinant variants. We also showed that the recombinant variants with replicative advantages were less sensitive to ribavirin in vitro. Finally, we found that the mechanisms leading to such a replicative advantage do not seem to rely on the post-translational modifications introduced by the human gene fragment that could have modified the function of the viral protein. The mechanisms involved in improving the replication of such recombinant viruses remain to be explored.
Keywords: hepatitis E virus; ribavirin; viral fitness; virus–host recombinant variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Kamar N, Selves J, Mansuy J-M, Ouezzani L, Péron J-M, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel J-P, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358:811–817. doi:10.1056/NEJMoa0706992 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

