Uncharacterized yeast gene YBR238C, an effector of TORC1 signaling in a mitochondrial feedback loop, accelerates cellular aging via HAP4- and RMD9-dependent mechanisms
- PMID: 38713053
- PMCID: PMC11076046
- DOI: 10.7554/eLife.92178
Uncharacterized yeast gene YBR238C, an effector of TORC1 signaling in a mitochondrial feedback loop, accelerates cellular aging via HAP4- and RMD9-dependent mechanisms
Abstract
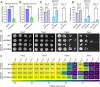
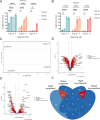
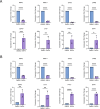
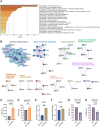
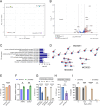
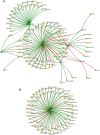
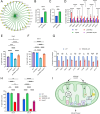
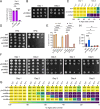
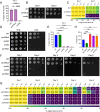
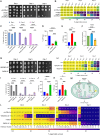
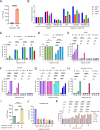
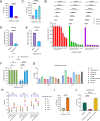
Uncovering the regulators of cellular aging will unravel the complexity of aging biology and identify potential therapeutic interventions to delay the onset and progress of chronic, aging-related diseases. In this work, we systematically compared genesets involved in regulating the lifespan of Saccharomyces cerevisiae (a powerful model organism to study the cellular aging of humans) and those with expression changes under rapamycin treatment. Among the functionally uncharacterized genes in the overlap set, YBR238C stood out as the only one downregulated by rapamycin and with an increased chronological and replicative lifespan upon deletion. We show that YBR238C and its paralog RMD9 oppositely affect mitochondria and aging. YBR238C deletion increases the cellular lifespan by enhancing mitochondrial function. Its overexpression accelerates cellular aging via mitochondrial dysfunction. We find that the phenotypic effect of YBR238C is largely explained by HAP4- and RMD9-dependent mechanisms. Furthermore, we find that genetic- or chemical-based induction of mitochondrial dysfunction increases TORC1 (Target of Rapamycin Complex 1) activity that, subsequently, accelerates cellular aging. Notably, TORC1 inhibition by rapamycin (or deletion of YBR238C) improves the shortened lifespan under these mitochondrial dysfunction conditions in yeast and human cells. The growth of mutant cells (a proxy of TORC1 activity) with enhanced mitochondrial function is sensitive to rapamycin whereas the growth of defective mitochondrial mutants is largely resistant to rapamycin compared to wild type. Our findings demonstrate a feedback loop between TORC1 and mitochondria (the
Keywords: S. cerevisiae; cellular aging; genetics; genomics; human; lifespan; mitochondria; nutrient signaling; target of rapamycin complex 1; uncharacterized genes.
© 2023, Alfatah et al.
Conflict of interest statement
MA, JL, YZ, AN, TC, SY, NF, JL, BE, FE No competing interests declared
Figures
Update of
- doi: 10.1101/2023.07.04.547743
- doi: 10.7554/eLife.92178.1
- doi: 10.7554/eLife.92178.2
References
-
- Alfatah M, Wong JH, Krishnan VG, Lee YC, Sin QF, Goh CJH, Kong KW, Lee WT, Lewis J, Hoon S, Arumugam P. TORC1 regulates the transcriptional response to glucose and developmental cycle via the Tap42-Sit4-Rrd1/2 pathway in Saccharomyces cerevisiae. BMC Biology. 2021;19:95. doi: 10.1186/s12915-021-01030-3. - DOI - PMC - PubMed
-
- Bauer JP, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin ML, Heijnen JJ, Hoare M, Lange HC, Madden EA, Niederberger P, Nielsen J, Parrou JL, Petit T, Porro D, Reuss M, Rizzi M, Steensma HY, Verrips CT, Vindeløv J, Pronk JT. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme and Microbial Technology. 2000;26:706–714. doi: 10.1016/s0141-0229(00)00162-9. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

