A conserved molecular logic for neurogenesis to gliogenesis switch in the cerebral cortex
- PMID: 38713624
- PMCID: PMC11098099
- DOI: 10.1073/pnas.2321711121
A conserved molecular logic for neurogenesis to gliogenesis switch in the cerebral cortex
Abstract
During development, neural stem cells in the cerebral cortex, also known as radial glial cells (RGCs), generate excitatory neurons, followed by production of cortical macroglia and inhibitory neurons that migrate to the olfactory bulb (OB). Understanding the mechanisms for this lineage switch is fundamental for unraveling how proper numbers of diverse neuronal and glial cell types are controlled. We and others recently showed that Sonic Hedgehog (Shh) signaling promotes the cortical RGC lineage switch to generate cortical oligodendrocytes and OB interneurons. During this process, cortical RGCs generate intermediate progenitor cells that express critical gliogenesis genes Ascl1, Egfr, and Olig2. The increased Ascl1 expression and appearance of Egfr+ and Olig2+ cortical progenitors are concurrent with the switch from excitatory neurogenesis to gliogenesis and OB interneuron neurogenesis in the cortex. While Shh signaling promotes Olig2 expression in the developing spinal cord, the exact mechanism for this transcriptional regulation is not known. Furthermore, the transcriptional regulation of Olig2 and Egfr has not been explored. Here, we show that in cortical progenitor cells, multiple regulatory programs, including Pax6 and Gli3, prevent precocious expression of Olig2, a gene essential for production of cortical oligodendrocytes and astrocytes. We identify multiple enhancers that control Olig2 expression in cortical progenitors and show that the mechanisms for regulating Olig2 expression are conserved between the mouse and human. Our study reveals evolutionarily conserved regulatory logic controlling the lineage switch of cortical neural stem cells.
Keywords: Olig2; enhancer; gliogenesis; lineage switch; neurogenesis.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
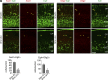
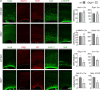
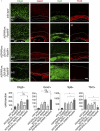

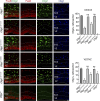
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

