E4BP4 in macrophages induces an anti-inflammatory phenotype that ameliorates the severity of colitis
- PMID: 38714733
- PMCID: PMC11076557
- DOI: 10.1038/s42003-024-06099-4
E4BP4 in macrophages induces an anti-inflammatory phenotype that ameliorates the severity of colitis
Abstract
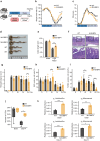
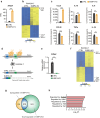
Macrophages are versatile cells of the innate immune system that work by altering their pro- or anti-inflammatory features. Their dysregulation leads to inflammatory disorders such as inflammatory bowel disease. We show that macrophage-specific upregulation of the clock output gene and transcription factor E4BP4 reduces the severity of colitis in mice. RNA-sequencing and single-cell analyses of macrophages revealed that increased expression of E4BP4 leads to an overall increase in expression of anti-inflammatory genes including Il4ra with a concomitant reduction in pro-inflammatory gene expression. In contrast, knockout of E4BP4 in macrophages leads to increased proinflammatory gene expression and decreased expression of anti-inflammatory genes. ChIP-seq and ATAC-seq analyses further identified Il4ra as a target of E4BP4, which drives anti-inflammatory polarization in macrophages. Together, these results reveal a critical role for E4BP4 in regulating macrophage inflammatory phenotypes and resolving inflammatory bowel diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

