Deregulated DNA ADP-ribosylation impairs telomere replication
- PMID: 38714889
- PMCID: PMC11102865
- DOI: 10.1038/s41594-024-01279-6
Deregulated DNA ADP-ribosylation impairs telomere replication
Abstract
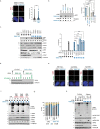
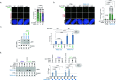
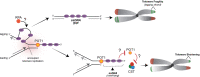
The recognition that DNA can be ADP ribosylated provides an unexpected regulatory level of how ADP-ribosylation contributes to genome stability, epigenetics and immunity. Yet, it remains unknown whether DNA ADP-ribosylation (DNA-ADPr) promotes genome stability and how it is regulated. Here, we show that telomeres are subject to DNA-ADPr catalyzed by PARP1 and removed by TARG1. Mechanistically, we show that DNA-ADPr is coupled to lagging telomere DNA strand synthesis, forming at single-stranded DNA present at unligated Okazaki fragments and on the 3' single-stranded telomere overhang. Persistent DNA-linked ADPr, due to TARG1 deficiency, eventually leads to telomere shortening. Furthermore, using the bacterial DNA ADP-ribosyl-transferase toxin to modify DNA at telomeres directly, we demonstrate that unhydrolyzed DNA-linked ADP-ribose compromises telomere replication and telomere integrity. Thus, by identifying telomeres as chromosomal targets of PARP1 and TARG1-regulated DNA-ADPr, whose deregulation compromises telomere replication and integrity, our study highlights and establishes the critical importance of controlling DNA-ADPr turnover for sustained genome stability.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

