Therapeutic efficacy of a K5-specific phage and depolymerase against Klebsiella pneumoniae in a mouse model of infection
- PMID: 38715095
- PMCID: PMC11077817
- DOI: 10.1186/s13567-024-01311-z
Therapeutic efficacy of a K5-specific phage and depolymerase against Klebsiella pneumoniae in a mouse model of infection
Abstract
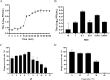
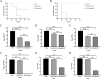
Klebsiella pneumoniae has become one of the most intractable gram-negative pathogens infecting humans and animals due to its severe antibiotic resistance. Bacteriophages and protein products derived from them are receiving increasing amounts of attention as potential alternatives to antibiotics. In this study, we isolated and investigated the characteristics of a new lytic phage, P1011, which lyses K5 K. pneumoniae specifically among 26 serotypes. The K5-specific capsular polysaccharide-degrading depolymerase dep1011 was identified and expressed. By establishing murine infection models using bovine strain B16 (capable of supporting phage proliferation) and human strain KP181 (incapable of sustaining phage expansion), we explored the safety and efficacy of phage and dep1011 treatments against K5 K. pneumoniae. Phage P1011 resulted in a 60% survival rate of the mice challenged with K. pneumoniae supporting phage multiplication, concurrently lowering the bacterial burden in their blood, liver, and lungs. Unexpectedly, even when confronted with bacteria impervious to phage multiplication, phage therapy markedly decreased the number of viable organisms. The protective efficacy of the depolymerase was significantly better than that of the phage. The depolymerase achieved 100% survival in both treatment groups regardless of phage propagation compatibility. These findings indicated that P1011 and dep1011 might be used as potential antibacterial agents to control K5 K. pneumoniae infection.
Keywords: Klebsiella pneumoniae; K5; capsule; depolymerase; hypervirulent; phage.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Two T7-like Bacteriophages, K5-2 and K5-4, Each Encodes Two Capsule Depolymerases: Isolation and Functional Characterization.Sci Rep. 2017 Jul 4;7(1):4624. doi: 10.1038/s41598-017-04644-2. Sci Rep. 2017. PMID: 28676686 Free PMC article.
-
Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment.J Infect Dis. 2014 Dec 1;210(11):1734-44. doi: 10.1093/infdis/jiu332. Epub 2014 Jul 7. J Infect Dis. 2014. PMID: 25001459
-
Targeted phage hunting to specific Klebsiella pneumoniae clinical isolates is an efficient antibiotic resistance and infection control strategy.Microbiol Spectr. 2024 Oct 3;12(10):e0025424. doi: 10.1128/spectrum.00254-24. Epub 2024 Aug 28. Microbiol Spectr. 2024. PMID: 39194291 Free PMC article.
-
The potential use of bacteriophages as antibacterial agents against Klebsiella pneumoniae.Virol J. 2024 Aug 19;21(1):191. doi: 10.1186/s12985-024-02450-7. Virol J. 2024. PMID: 39160541 Free PMC article. Review.
-
Klebsiella pneumoniae infections and phage therapy.Indian J Med Microbiol. 2024 Nov-Dec;52:100736. doi: 10.1016/j.ijmmb.2024.100736. Epub 2024 Oct 5. Indian J Med Microbiol. 2024. PMID: 39357832 Review.
Cited by
-
Specificity and diversity of Klebsiella pneumoniae phage-encoded capsule depolymerases.Essays Biochem. 2024 Dec 17;68(5):661-677. doi: 10.1042/EBC20240015. Essays Biochem. 2024. PMID: 39668555 Free PMC article. Review.
-
Phage therapy for Klebsiella pneumoniae: Understanding bacteria-phage interactions for therapeutic innovations.PLoS Pathog. 2025 Apr 8;21(4):e1012971. doi: 10.1371/journal.ppat.1012971. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40198880 Free PMC article. Review.
-
Phage-Based Therapy in Combination with Antibiotics: A Promising Alternative against Multidrug-Resistant Gram-Negative Pathogens.Pathogens. 2024 Oct 14;13(10):896. doi: 10.3390/pathogens13100896. Pathogens. 2024. PMID: 39452768 Free PMC article. Review.
-
Phage Therapy: A Promising Treatment Strategy against Infections Caused by Multidrug-resistant Klebsiella pneumoniae.Curr Pharm Des. 2025;31(13):1007-1019. doi: 10.2174/0113816128343976241117183624. Curr Pharm Des. 2025. PMID: 39757682 Review.
-
Identification and preclinical efficacy evaluation of two lytic bacteriophages targeting highly virulent and multidrug-resistant Klebsiella pneumoniae.Ann Clin Microbiol Antimicrob. 2025 Aug 20;24(1):46. doi: 10.1186/s12941-025-00812-9. Ann Clin Microbiol Antimicrob. 2025. PMID: 40835923 Free PMC article.
References
-
- Uzairue LI, Rabaan AA, Adewumi FA, Okolie OJ, Folorunso JB, Bakhrebah MA, Garout M, Alfouzan WA, Halwani MA, Alamri AA, Halawani SA, Alshahrani FS, Hasan A, Mutair AA, Alhumaid S, Etafo J, Utip I, Odoh IM, Uwaezuoke NS. Global prevalence of colistin resistance in Klebsiella pneumoniae from bloodstream infection: a systematic review and meta-analysis. Pathogens. 2022;11:1092. doi: 10.3390/pathogens11101092. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- SCKJ-JYRC-2022-80/Sanya Yazhou Bay Science and Technology City
- NAUSY-MS12/The Guidance Foundation, the Sanya Institute of Nanjing Agricultural University
- KYCX23_0829/Graduate Research and Innovation Projects of Jiangsu Province
- EM202303/Molecular typing and bacteriophage control of pathogenic bacteria in enterobacteria
- 82002108/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Research Materials

