A practical guide to the implementation of artificial intelligence in orthopaedic research-Part 2: A technical introduction
- PMID: 38715910
- PMCID: PMC11076014
- DOI: 10.1002/jeo2.12025
A practical guide to the implementation of artificial intelligence in orthopaedic research-Part 2: A technical introduction
Abstract
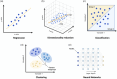
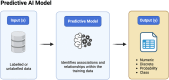
Recent advances in artificial intelligence (AI) present a broad range of possibilities in medical research. However, orthopaedic researchers aiming to participate in research projects implementing AI-based techniques require a sound understanding of the technical fundamentals of this rapidly developing field. Initial sections of this technical primer provide an overview of the general and the more detailed taxonomy of AI methods. Researchers are presented with the technical basics of the most frequently performed machine learning (ML) tasks, such as classification, regression, clustering and dimensionality reduction. Additionally, the spectrum of supervision in ML including the domains of supervised, unsupervised, semisupervised and self-supervised learning will be explored. Recent advances in neural networks (NNs) and deep learning (DL) architectures have rendered them essential tools for the analysis of complex medical data, which warrants a rudimentary technical introduction to orthopaedic researchers. Furthermore, the capability of natural language processing (NLP) to interpret patterns in human language is discussed and may offer several potential applications in medical text classification, patient sentiment analysis and clinical decision support. The technical discussion concludes with the transformative potential of generative AI and large language models (LLMs) on AI research. Consequently, this second article of the series aims to equip orthopaedic researchers with the fundamental technical knowledge required to engage in interdisciplinary collaboration in AI-driven orthopaedic research.
Level of evidence: Level IV.
Keywords: artificial intelligence; machine learning; orthopaedics; research methods; sports medicine.
© 2024 The Author(s). Journal of Experimental Orthopaedics published by John Wiley & Sons Ltd on behalf of European Society of Sports Traumatology, Knee Surgery and Arthroscopy.
Conflict of interest statement
Michael T. Hirschmann is a consultant for Medacta, Symbios and Depuy Synthes. Kristian Samuelsson is a member on the board of directors for Getinge AB (publ). Robert Feldt is Chief Technology Officer and founder in Accelerandium AB, a software consultancy company.
Figures


References
-
- Amatriain, X. (2023) Transformer models: an introduction and catalog. To be published in arXiv. [Preprint] Available from: 10.48550/arXiv.2302.07730 - DOI
-
- An, G. , Akiba, M. , Omodaka, K. , Nakazawa, T. & Yokota, H. (2021) Hierarchical deep learning models using transfer learning for disease detection and classification based on small number of medical images. Scientific Reports, 11, 4250. Available from: 10.1038/s41598-021-83503-7 - DOI - PMC - PubMed
-
- Anandika, A. , Mishra, S.P. & Das, M. (2021) Review on usage of hidden markov model in natural language processing. In: Intelligent and Cloud Computing: Proceedings of ICICC 2019, vol. 1, Singapore: Springer, pp. 415–423. Available from: 10.1007/978-981-15-5971-6_45 - DOI
-
- Awad, M. & Khanna, R. (2015) Support vector machines for classification. In: Awad, M. & Khanna, R. (Eds.) Efficient learning machines: theories, concepts, and applications for engineers and system designers. Berkeley, CA: Apress, pp. 39–66. Available from: 10.1007/978-1-4302-5990-9_3 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

