Comparison of qPCR and metabarcoding for environmental DNA surveillance of a freshwater parasite
- PMID: 38716167
- PMCID: PMC11074384
- DOI: 10.1002/ece3.11382
Comparison of qPCR and metabarcoding for environmental DNA surveillance of a freshwater parasite
Abstract
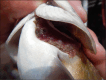
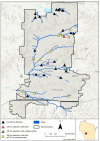
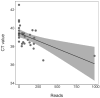
Analysis of environmental DNA (eDNA) has been successfully used across freshwater ecological parasitology to inform management of ecologically and economically important species. However, most studies have used species-specific quantitative polymerase chain reaction (qPCR) assays to detect target taxa. While generally effective, this approach limits the amount of community and management-supporting data that can be obtained from eDNA samples. If eDNA metabarcoding could be conducted with the same accuracy of a single species approach, researchers could simultaneously detect a target species while obtaining vast community data from eDNA samples. We sampled 38 freshwater sites on Fort McCoy, Wisconsin and compared qPCR to metabarcoding for eDNA detection of the ectoparasitic gill louse Salmincola edwardsii, an obligate parasite of Salvelinus fishes (chars). We found no evidence to suggest S. edwardsii occupancy or detection probabilities differed between qPCR and metabarcoding. Further, we found that the number of S. edwardsii reads from metabarcoding were negatively predictive of C T values from qPCR (C T value indicates cycle a significant amount of target eDNA is detected, with lower C Ts indicative of more DNA), demonstrating that our metabarcoding reads positively predicted qPCR DNA quantities. However, the number of reads was not predictive of overall qPCR score (number of positive qPCR replicates). In addition to S. edwardsii, metabarcoding led to the detection of a vast community of over 2600 invertebrate taxa. We underscore the necessity for conducting similar analyses across environments and target species, as the ecology of eDNA will vary on a per-study basis. Our results suggest that eDNA metabarcoding provides a highly sensitive and accurate method for detecting parasitic gill lice while also illuminating the broader biological community and co-occurrence of species in the environment.
Keywords: Salmincola edwardsii; eDNA; metabarcoding; occupancy modeling; qPCR.
© 2024 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Andres, K. J. , Lambert, T. D. , Lodge, D. M. , Andres, J. , & Jackson, J. R. (2022). Combining sampling gear to optimally inventory species highlights the efficiency of eDNA metabarcoding. Environmental DNA, 5, 146–157. 10.1002/edn3.366 - DOI
-
- Bakker, J. , Wangensteen, O. S. , Champman, D. D. , Boussarie, G. , Buddo, D. , Guttridge, T. L. , Hertler, H. , Mouillot, D. , Vigliola, L. , & Mariani, S. (2017). Environmental DNA reveals tropical shark diversity in contrasting levels of anthropogenic impact. Scientific Reports, 7, 16886. 10.1038/s41598-017-17150-2 - DOI - PMC - PubMed
-
- Barber, I. , Hoare, D. , & Krause, J. (2000). Effects of parasites on fish behavior: A review and evolutionary perspective. Reviews in Fish Biology and Fisheries, 10, 131–165. 10.1023/A:1016658224470 - DOI
-
- Barnes, M. A. , & Turner, C. R. (2016). The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics, 17, 1–17. 10.1007/s10592-015-0775-4 - DOI
LinkOut - more resources
Full Text Sources

