D3S-001, a KRAS G12C Inhibitor with Rapid Target Engagement Kinetics, Overcomes Nucleotide Cycling, and Demonstrates Robust Preclinical and Clinical Activities
- PMID: 38717075
- PMCID: PMC11372373
- DOI: 10.1158/2159-8290.CD-24-0006
D3S-001, a KRAS G12C Inhibitor with Rapid Target Engagement Kinetics, Overcomes Nucleotide Cycling, and Demonstrates Robust Preclinical and Clinical Activities
Abstract
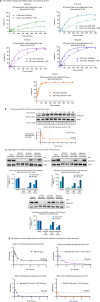
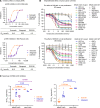
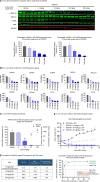
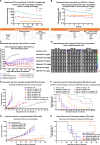
First-generation KRAS G12C inhibitors, such as sotorasib and adagrasib, are limited by the depth and duration of clinical responses. One potential explanation for their modest clinical activity is the dynamic "cycling" of KRAS between its guanosine diphosphate (GDP)- and guanosine triphosphate (GTP)-bound states, raising controversy about whether targeting the GDP-bound form can fully block this oncogenic driver. We herein report that D3S-001, a next-generation GDP-bound G12C inhibitor with faster target engagement (TE) kinetics, depletes cellular active KRAS G12C at nanomolar concentrations. In the presence of growth factors, such as epithelial growth factor and hepatocyte growth factor, the ability of sotorasib and adagrasib to inhibit KRAS was compromised whereas the TE kinetics of D3S-001 was nearly unaffected, a unique feature differentiating D3S-001 from other GDP-bound G12C inhibitors. Furthermore, the high covalent potency and cellular TE efficiency of D3S-001 contributed to robust antitumor activity preclinically and translated into promising clinical efficacy in an ongoing phase 1 trial (NCT05410145). Significance: The kinetic study presented in this work unveils, for the first time, that a GDP-bound conformation-selective KRAS G12C inhibitor can potentially deplete cellular active KRAS in the presence of growth factors and offers new insights into the critical features that drive preclinical and clinical efficacy for this class of drugs.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
C. Chen reports other support from D3 Bio outside the submitted work. H. Rui reports other support from D3 Bio outside the submitted work. S.M. Lim reports grants from AstraZeneca, Hutchison, and Roche and personal fees from AstraZeneca, Boehringer Ingelheim, Hutchison, Roche, Simcere, and Innovent Biologics outside the submitted work. T. Mok reports personal fees from Abbvie Inc., ACEA Pharma, and Adagene; personal fees and other support from Alentis Therapeutics AG; personal fees from Alpha Biopharma Co. Ltd., Amgen, Amoy Diagnostics Co. Ltd., AnHeart Therapeutics Inc., AVEO Pharmaceuticals Inc., Bayer HealthCare, BeiGene, BerGenBio ASA, Berry Oncology, Boehringer Ingelheim, and Blueprint Medicines Corporation; grants and personal fees from Bristol Myers Squibb; personal fees from Bowtie Life Insurance Co Ltd., Bridge Biotherapeutics Inc., C4 Therapeutics Inc., Cirina Ltd., Covidien LP, CStone Pharmaceuticals, and Curio Science; personal fees and other support from D3 Bio Ltd.; personal fees from Da Volterra, Daiichi Sankyo Inc., Daz Group, Eisai, Elevation Oncology, and F. Hoffmann-La Roche Ltd.; personal fees from Fishawack Facilitate Ltd.; grants and personal fees from G1 Therapeutics Inc.; personal fees from geneDecode Co. Ltd, Genentech, Gilead Sciences Inc., GLG’s Healthcare, Gritstone Oncology Inc., Guardant Health, Hengrui Therapeutics Inc., and HiberCell Inc.; personal fees, nonfinancial support, and other support from HutchMed; personal fees from Ignyta Inc., Illumina Inc., Imagene AI Ltd., Gritstone Oncology Inc., Incyte Corporation, Inivata, InMed Medical Communication, IQVIA, Janssen, Janssen Pharmaceutica NV, Jiahui Holdings Co. Limited, and Lakeshore Biotech; personal fees and nonfinancial support from LiangYiHui Healthcare; personal fees from Lilly, Loxo-Oncology, and Lucence Health Inc.; personal fees and other support from Lunit Inc.; personal fees from MD Health Brazil, Medscape LLC, Medtronic, and Merck Pharmaceuticals HK Ltd.; grants and personal fees from Merck Serono; grants, personal fees, and nonfinancial support from Merck Sharp & Dohme; personal fees from Mirati Therapeutics Inc.; personal fees and nonfinancial support from MiRXES; personal fees from MoreHealth; grants, personal fees, and nonfinancial support from Novartis; personal fees from Novocure GmbH, Omega Therapeutics Inc., OrigiMed Co. Ltd., OSE Immunotherapeutics, P. Permanyer SL, and PeerVoice; grants, personal fees, and nonfinancial support from Pfizer; personal fees from Phanes Therapeutics; personal fees and nonfinancial support from Physicians’ Education Resource; personal fees and other support from Prenetics Global Limited; personal fees from PrIME Oncology, Puma Biotechnology Inc., Qiming Development (HK) Ltd., Regeneron Pharmaceuticals Inc., and Research to Practice; grants, personal fees, and nonfinancial support from Roche Pharmaceuticals/Diagnostics/Foundation One; personal fees from Sanofi-Aventis, Schrödinger Inc., and Seagen International GmbH; grants and personal fees from SFJ Pharmaceutical; personal fees from Shanghai BeBirds Translation & Consulting Co. Ltd., Shanghai Promedican Pharmaceuticals Co. Ltd., Simcere of America Inc, Simcere Zaiming Inc., Summit Therapeutics Sub Inc., Synergy Research, and Taiho Pharmaceutical Co. Ltd; grants and personal fees from Takeda; personal fees from Tigermed, Touch Independent Medical Education Ltd, Vertex Pharmaceuticals, Virtus Medical Group, XENCOR Inc., and Yuhan Corporation; personal fees and nonfinancial support from Zai Lab; grants, personal fees, nonfinancial support, and other support from AstraZeneca PLC; personal fees, nonfinancial support, and other support from HutchMed; personal fees and other support from Aurora; personal fees and other support from Insighta; other support from Yinson Capital Pte. Ltd.; personal fees from The Chinese University of Hong Kong; and grants from XCovery outside the submitted work. B.C. Cho reports grants from MOGAM Institute, LG Chem, Oscotec, Interpark Bio Convergence Corp, GIInnovation, GI-Cell, Abion, Abbvie, AstraZeneca, Bayer, Blueprint Medicines, Boehringer Ingelheim, Champions Oncology, CJ bioscience, CJ Blossom Park, Cyrus, Dizal Pharma, Genexine, Janssen, Lilly, MSD, Novartis, Nuvalent, Oncternal, Ono, Regeneron, Dong-A ST, Bridgebio therapeutics, Yuhan, ImmuneOncia, Illumina, KANAPH therapeutics, Therapex, JINTSbio, Hanmi, CHA Bundang Medical Center, Vertical Bio AG, personal fees from Abion, BeiGene, Novartis, AstraZeneca, Boehringer Ingelheim, Roche, BMS, CJ, CureLogen, Cyrus Therapeutics, Ono, Onegene Biotechnology, Yuhan, Pfizer, Eli Lilly, GI-Cell, Guardant, HK Inno-N, Imnewrun Biosciences Inc., Janssen, Takeda, MSD, Janssen, Medpacto, Blueprint medicines, RandBio, and Hanmi; personal fees from KANAPH Therapeutic Inc, Bridgebio therapeutics, Cyrus Therapeutics, Guardant Health, Oscotec Inc, J INTS Bio, Therapex Co., Ltd, Gliead, and Amgen; personal fees from TheraCanVac Inc, Gencurix Inc, Bridgebio therapeutics, KANAPH Therapeutic Inc, Cyrus therapeutics, Interpark Bio Convergence Corp., and J INTS BIO; personal fees from J INTS BIO, Champions Oncology, Crown Bioscience, Imagen, and PearlRiver Bio GmbH; and other support from DAAN Biotherapeutics outside the submitted work. No disclosures were reported by the other authors.
Figures


References
-
- Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRASG12C somatic mutations across race, sex, and cancer type. N Engl J Med 2021;384:185–7. - PubMed
-
- Sebastian M, Eberhardt WEE, Hoffknecht P, Metzenmacher M, Wehler T, Kokowski K, et al. . KRAS G12C-mutated advanced non-small cell lung cancer: a real-world cohort from the German prospective, observational, nation-wide CRISP registry (AIO-TRK-0315). Lung Cancer 2021;154:51–61. - PubMed
-
- Salem M, El-Refai S, Sha W, Grothey A, Puccini A, George T, et al. . O-3 Characterization of KRAS mutation variants and prevalence of KRAS-G12C in gastrointestinal malignancies. Ann Oncol 2021;32:S218.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

