Fecal microbiota transplantation: current challenges and future landscapes
- PMID: 38717124
- PMCID: PMC11325845
- DOI: 10.1128/cmr.00060-22
Fecal microbiota transplantation: current challenges and future landscapes
Abstract
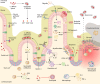
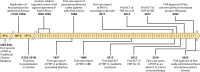
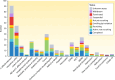
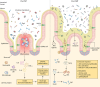
SUMMARYGiven the importance of gut microbial homeostasis in maintaining health, there has been considerable interest in developing innovative therapeutic strategies for restoring gut microbiota. One such approach, fecal microbiota transplantation (FMT), is the main "whole gut microbiome replacement" strategy and has been integrated into clinical practice guidelines for treating recurrent Clostridioides difficile infection (rCDI). Furthermore, the potential application of FMT in other indications such as inflammatory bowel disease (IBD), metabolic syndrome, and solid tumor malignancies is an area of intense interest and active research. However, the complex and variable nature of FMT makes it challenging to address its precise functionality and to assess clinical efficacy and safety in different disease contexts. In this review, we outline clinical applications, efficacy, durability, and safety of FMT and provide a comprehensive assessment of its procedural and administration aspects. The clinical applications of FMT in children and cancer immunotherapy are also described. We focus on data from human studies in IBD in contrast with rCDI to delineate the putative mechanisms of this treatment in IBD as a model, including colonization resistance and functional restoration through bacterial engraftment, modulating effects of virome/phageome, gut metabolome and host interactions, and immunoregulatory actions of FMT. Furthermore, we comprehensively review omics technologies, metagenomic approaches, and bioinformatics pipelines to characterize complex microbial communities and discuss their limitations. FMT regulatory challenges, ethical considerations, and pharmacomicrobiomics are also highlighted to shed light on future development of tailored microbiome-based therapeutics.
Keywords: Clostridioides difficile infection; donor screening; fecal microbiota transplantation; human microbiome; microbial engraftment.
Conflict of interest statement
D.K. has received consulting fees from Ferring; E.M.T. and E.J.K. received an unrestricted research grant from Vedanta Biosciences; T.M.M. has received consulting fees from Takeda Pharmaceuticals and served as an advisor for CHAIN Biotechnology.
Figures


References
-
- Burcham ZM, Garneau NL, Comstock SS, Tucker RM, Knight R, Metcalf JL, Miranda A, Reinhart B, Meyers D, Woltkamp D, Boxer E, Hutchens J, Kim K, Archer M, McAteer M, Huss P, Defonseka R, Stahle S, Babu S, Nuessle T, Schowinsky V, Covert W, Truman W, Reusser W, Taste Lab Citizen S. 2020. Patterns of oral microbiota diversity in adults and children: a crowdsourced population study. Sci Rep 10:2133. doi:10.1038/s41598-020-59016-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

