LYVE-1-expressing Macrophages Modulate the Hyaluronan-containing Extracellular Matrix in the Mammary Stroma and Contribute to Mammary Tumor Growth
- PMID: 38717149
- PMCID: PMC11141485
- DOI: 10.1158/2767-9764.CRC-24-0205
LYVE-1-expressing Macrophages Modulate the Hyaluronan-containing Extracellular Matrix in the Mammary Stroma and Contribute to Mammary Tumor Growth
Abstract
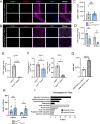
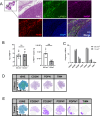
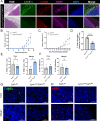
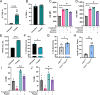
Macrophages represent a heterogeneous myeloid population with diverse functions in normal tissues and tumors. While macrophages expressing the cell surface marker lymphatic vessel endothelial hyaluronan receptor 1 (LYVE-1) have been identified in stromal regions of the normal mammary gland and in the peritumoral stroma, their functions within these regions are not well understood. Using a genetic mouse model of LYVE-1+ macrophage depletion, we demonstrate that loss of LYVE-1+ macrophages is associated with altered extracellular matrix remodeling in the normal mammary gland and reduced mammary tumor growth in vivo. In further studies focused on investigating the functions of LYVE-1+ macrophages in the tumor microenvironment, we demonstrate that LYVE-1 expression correlates with an increased ability of macrophages to bind, internalize, and degrade hyaluronan. Consistent with this, we show that depletion of LYVE-1+ macrophages correlates with increased hyaluronan accumulation in both the normal mammary gland and in mammary tumors. Analysis of single-cell RNA sequencing of macrophages isolated from these tumors reveals that depletion of LYVE-1+ macrophages in tumors drives a shift in the majority of the remaining macrophages toward a proinflammatory phenotype, as well as an increase in CD8+ T-cell infiltration. Together, these findings indicate that LYVE-1+ macrophages represent a tumor-promoting anti-inflammatory subset of macrophages that contributes to hyaluronan remodeling in the tumor microenvironment.
Significance: We have identified a macrophage subset in mouse mammary tumors associated with tumor structural components. When this macrophage subset is absent in tumors, we report a delay in tumor growth and an increase in antitumor immune cells. Understanding the functions of distinct macrophage subsets may allow for improved therapeutic strategies for patients with breast cancer.
© 2024 The Authors; Published by the American Association for Cancer Research.
Figures

References
-
- Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17–producing T cell responses. Nat Immunol 2007;8:1086–94. - PubMed
-
- Hagemann T, Robinson SC, Schulz M, Trümper L, Balkwill FR, Binder C. Enhanced invasiveness of breast cancer cell lines upon co-cultivation with macrophages is due to TNF-α dependent up-regulation of matrix metalloproteases. Carcinogenesis 2004;25:1543–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

