Senataxin deficiency disrupts proteostasis through nucleolar ncRNA-driven protein aggregation
- PMID: 38717338
- PMCID: PMC11080644
- DOI: 10.1083/jcb.202309036
Senataxin deficiency disrupts proteostasis through nucleolar ncRNA-driven protein aggregation
Abstract
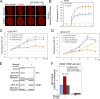
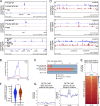
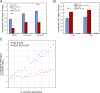
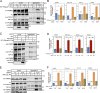
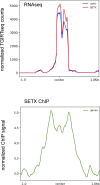
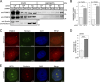
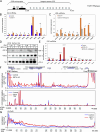
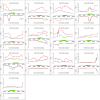
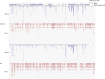
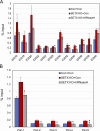
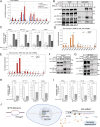
Senataxin is an evolutionarily conserved RNA-DNA helicase involved in DNA repair and transcription termination that is associated with human neurodegenerative disorders. Here, we investigated whether Senataxin loss affects protein homeostasis based on previous work showing R-loop-driven accumulation of DNA damage and protein aggregates in human cells. We find that Senataxin loss results in the accumulation of insoluble proteins, including many factors known to be prone to aggregation in neurodegenerative disorders. These aggregates are located primarily in the nucleolus and are promoted by upregulation of non-coding RNAs expressed from the intergenic spacer region of ribosomal DNA. We also map sites of R-loop accumulation in human cells lacking Senataxin and find higher RNA-DNA hybrids within the ribosomal DNA, peri-centromeric regions, and other intergenic sites but not at annotated protein-coding genes. These findings indicate that Senataxin loss affects the solubility of the proteome through the regulation of transcription-dependent lesions in the nucleus and the nucleolus.
© 2024 Wen et al.
Conflict of interest statement
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. A.M. Lambowitz reported multiple patents and patent applications on TGIRT-seq enzymes and methods licensed “Dana Farber Genomic Services core facility.” No other disclosures were reported.
Figures






References
-
- Airoldi, G., Guidarelli A., Cantoni O., Panzeri C., Vantaggiato C., Bonato S., Grazia D’Angelo M., Falcone S., De Palma C., Tonelli A., et al. . 2010. Characterization of two novel SETX mutations in AOA2 patients reveals aspects of the pathophysiological role of senataxin. Neurogenetics. 11:91–100. 10.1007/s10048-009-0206-0 - DOI - PubMed
-
- Alzu, A., Bermejo R., Begnis M., Lucca C., Piccini D., Carotenuto W., Saponaro M., Brambati A., Cocito A., Foiani M., and Liberi G.. 2012. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell. 151:835–846. 10.1016/j.cell.2012.09.041 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

