Fluorescent Chiral Quantum Dots to Unveil Origin-Dependent Exosome Uptake and Cargo Release
- PMID: 38717870
- PMCID: PMC11393810
- DOI: 10.1021/acsabm.4c00296
Fluorescent Chiral Quantum Dots to Unveil Origin-Dependent Exosome Uptake and Cargo Release
Abstract
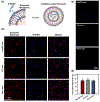
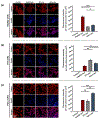
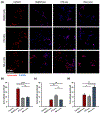
Exosomes are promising nanocarriers for drug delivery. Yet, it is challenging to apply exosomes in clinical use due to the limited understanding of their physiological functions. While cellular uptake of exosomes is generally known through endocytosis and/or membrane fusion, the mechanisms of origin-dependent cellular uptake and subsequent cargo release of exosomes into recipient cells are still unclear. Herein, we investigated the intricate mechanisms of exosome entry into recipient cells and intracellular cargo release. In this study, we utilized chiral graphene quantum dots (GQDs) as representatives of exosomal cargo, taking advantage of the superior permeability of chiral GQDs into lipid membranes as well as their excellent optical properties for tracking analysis. We observed that the preferential cellular uptake of exosomes derived from the same cell-of-origin (intraspecies exosomes) is higher than that of exosomes derived from different cell-of-origin (cross-species exosomes). This uptake enhancement was attributed to receptor-ligand interaction-mediated endocytosis, as we identified the expression of specific ligands on exosomes that favorably interact with their parental cells and confirmed the higher lysosomal entrapment of intraspecies exosomes (intraspecies endocytic uptake). On the other hand, we found that the uptake of cross-species exosomes primarily occurred through membrane fusion, followed by direct cargo release into the cytosol (cross-species direct fusion uptake). We revealed the underlying mechanisms involved in the cellular uptake and subsequent cargo release of exosomes depending on their cell-of-origin and recipient cell types. Overall, this study envisions valuable insights into further advancements in effective drug delivery using exosomes, as well as a comprehensive understanding of cellular communication, including disease pathogenesis.
Keywords: cell-of-origin; chirality; endocytosis; exosomes; graphene quantum dots; membrane fusion.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





Update of
-
Fluorescent Chiral Quantum Dots to Unveil Origin-Dependent Exosome Uptake and Cargo Release.bioRxiv [Preprint]. 2023 Dec 21:2023.12.20.572689. doi: 10.1101/2023.12.20.572689. bioRxiv. 2023. Update in: ACS Appl Bio Mater. 2024 May 20;7(5):3358-3374. doi: 10.1021/acsabm.4c00296. PMID: 38187632 Free PMC article. Updated. Preprint.
References
-
- Johnston J; Stone T; Wang Y Biomaterial-enabled 3D cell culture technologies for extracellular vesicle manufacturing. Biomater. Sci 2023, 11 (12), 4055. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
